Abstract
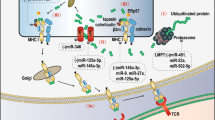
Cancer immunotherapy has revolutionized oncology practice. However, current protein and cell therapy tools used in cancer immunotherapy are far from perfect, and there is room for improvement regarding their efficacy and safety. RNA-based structures have diverse functions, ranging from gene expression and gene regulation to pro-inflammatory effects and the ability to specifically bind different molecules. These functions make them versatile tools that may advance cancer vaccines and immunomodulation, surpassing existing approaches. These technologies should not be considered as competitors of current immunotherapies but as partners in synergistic combinations and as a clear opportunity to reach more efficient and personalized results. RNA and RNA derivatives can be exploited therapeutically as a platform to encode protein sequences, provide innate pro-inflammatory signals to the immune system (such as those denoting viral infection), control the expression of other RNAs (including key immunosuppressive factors) post-transcriptionally and conform structural scaffoldings binding proteins that control immune cells by modifying their function. Nascent RNA immunotherapeutics include RNA vaccines encoding cancer neoantigens, mRNAs encoding immunomodulatory factors, viral RNA analogues, interference RNAs and protein-binding RNA aptamers. These approaches are already in early clinical development with promising safety and efficacy results.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
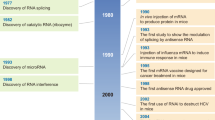
Gilbert, W. Origin of life: the RNA world. Nature 319, 618 (1986).
Pearce, B. K. D., Pudritz, R. E., Semenov, D. A. & Henning, T. K. Origin of the RNA world: the fate of nucleobases in warm little ponds. Proc. Natl Acad. Sci. USA 114, 11327–11332 (2017).
Roeder, R. G. & Rutter, W. J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature 224, 234–237 (1969).
Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261 (2015).
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47 (2016).
Kung, J. T., Colognori, D. & Lee, J. T. Long noncoding RNAs: past, present, and future. Genetics 193, 651–669 (2013).
Sainsbury, S., Bernecky, C. & Cramer, P. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 (2015).
Kwak, H. & Lis, J. T. Control of transcriptional elongation. Annu. Rev. Genet. 47, 483–508 (2013).
Richard, P. & Manley, J. L. Transcription termination by nuclear RNA polymerases. Genes Dev. 23, 1247–1269 (2009).
Ramanathan, A., Robb, G. B. & Chan, S. H. mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526 (2016).
Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18, 655–670 (2017).
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008).
Wojtowicz, W. M., Flanagan, J. J., Millard, S. S., Zipursky, S. L. & Clemens, J. C. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell 118, 619–633 (2004).
Di Giammartino, D. C., Nishida, K. & Manley, J. L. Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866 (2011).
Tian, B. & Manley, J. L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18, 18–30 (2017).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Kariko, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008). This article demonstrates that the incorporation of pseudouridine into mRNA leads to deimmunized RNAs with increased stability and translational capacity.
Hoernes, T. P. et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 44, 852–862 (2016).
Kozak, M. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115, 887–903 (1991).
Shi, Z. & Barna, M. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 31, 31–54 (2015).
Yang, F. & Schoenberg, D. R. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol. Cell 14, 435–445 (2004).
Conti, E. & Izaurralde, E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17, 316–325 (2005).
Kertesz, M. et al. Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107 (2010).
Lu, Z. et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165, 1267–1279 (2016).
Kruger, K. et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157 (1982).
Wochner, A., Attwater, J., Coulson, A. & Holliger, P. Ribozyme-catalyzed transcription of an active ribozyme. Science 332, 209–212 (2011).
Fitzgerald, K., Kallend, D. & Simon, A. A. Highly durable RNAi therapeutic inhibitor of PCSK9. N. Engl. J. Med. 376, e38 (2017).
Rand, T. A., Ginalski, K., Grishin, N. V. & Wang, X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl Acad. Sci. USA 101, 14385–14389 (2004).
Shi, M. et al. Redefining the invertebrate RNA virosphere. Nature 540, 539–543 (2016).
Koonin, E. V., Dolja, V. V. & Krupovic, M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480, 2–25 (2015).
Ranzani, M. et al. Lentiviral vector–based insertional mutagenesis identifies genes associated with liver cancer. Nat. Methods 10, 155 (2013).
Lauring, A. S., Frydman, J. & Andino, R. The role of mutational robustness in RNA virus evolution. Nat. Rev. Microbiol. 11, 327–336 (2013).
Schlee, M. & Hartmann, G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16, 566–580 (2016).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Matsumoto, M., Kikkawa, S., Kohase, M., Miyake, K. & Seya, T. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem. Biophys. Res. Commun. 293, 1364–1369 (2002).
Judge, A. D. et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 23, 457–462 (2005).
Ablasser, A. et al. Selection of molecular structure and delivery of RNA oligonucleotides to activate TLR7 versus TLR8 and to induce high amounts of IL-12p70 in primary human monocytes. J. Immunol. 182, 6824–6833 (2009).
Pichlmair, A. et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314, 997–1001 (2006).
Kato, H. et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205, 1601–1610 (2008).
Runge, S. et al. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLOS Pathog. 10, e1004081 (2014).
Sato, M. et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13, 539–548 (2000).
Tailor, P. et al. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity 27, 228–239 (2007).
Hervas-Stubbs, S. et al. Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 17, 2619–2627 (2011).
Bianchi, F., Pretto, S., Tagliabue, E., Balsari, A. & Sfondrini, L. Exploiting poly(I:C) to induce cancer cell apoptosis. Cancer Biol. Ther. 18, 747–756 (2017).
Salazar, A. M., Erlich, R. B., Mark, A., Bhardwaj, N. & Herberman, R. B. Therapeutic in situ autovaccination against solid cancers with intratumoral poly-ICLC: case report, hypothesis, and clinical trial. Cancer Immunol. Res. 2, 720–724 (2014).
Caskey, M. et al. Synthetic double-stranded RNA induces innate immune responses similar to a live viral vaccine in humans. J. Exp. Med. 208, 2357–2366 (2011).
Rodriguez-Ruiz, M. E. et al. Combined immunotherapy encompassing intratumoral poly-ICLC, dendritic-cell vaccination and radiotherapy in advanced cancer patients. Ann. Oncol. 29, 1312–1319 (2018). This clinical trial illustrates the activation of immune responses in patients with cancer who were treated with poly-ICLC, radiotherapy and DC vaccination.
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03262103 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02423863 (2015).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT01976585 (2013).
Tormo, D. et al. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell 16, 103–114 (2009).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02828098 (2016).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
De Beuckelaer, A., Grooten, J. & De Koker, S. Type I interferons modulate CD8(+) T cell immunity to mRNA vaccines. Trends Mol. Med. 23, 216–226 (2017).
Fioravanti, J. et al. Anchoring interferon alpha to apolipoprotein A-I reduces hematological toxicity while enhancing immunostimulatory properties. Hepatology 53, 1864–1873 (2011).
Gil, M. P. et al. Regulating type 1 IFN effects in CD8 T cells during viral infections: changing STAT4 and STAT1 expression for function. Blood 120, 3718–3728 (2012).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Kormann, M. S. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 (2011).
Andries, O. et al. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control Release 217, 337–344 (2015).
Kariko, K., Muramatsu, H., Ludwig, J. & Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 39, e142 (2011).
Thess, A. et al. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 23, 1456–1464 (2015).
Kauffman, K. J. et al. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 109, 78–87 (2016).
Judge, A. D., Bola, G., Lee, A. C. & MacLachlan, I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 13, 494–505 (2006).
Broering, R. et al. Chemical modifications on siRNAs avoid Toll-like-receptor-mediated activation of the hepatic immune system in vivo and in vitro. Int. Immunol. 26, 35–46 (2014).
Pardi, N., Hogan, M. J., Porter, F. W. & Weissman, D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018).
McConkey, S. J. et al. Enhanced T cell immunogenicity of plasmid DNA vaccines boosted by recombinant modified vaccinia virus Ankara in humans. Nat. Med. 9, 729–735 (2003).
Melero, I. et al. Therapeutic vaccines for cancer: an overview of clinical trials. Nat. Rev. Clin. Oncol. 11, 509–524 (2014).
Conry, R. M. et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 55, 1397–1400 (1995).
Boczkowski, D., Nair, S. K., Snyder, D. & Gilboa, E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J. Exp. Med. 184, 465–472 (1996).
Morse, M. A. et al. The feasibility and safety of immunotherapy with dendritic cells loaded with CEA mRNA following neoadjuvant chemoradiotherapy and resection of pancreatic cancer. Int. J. Gastrointest. Cancer 32, 1–6 (2002).
Heiser, A. et al. Autologous dendritic cells transfected with prostate-specific antigen RNA stimulate CTL responses against metastatic prostate tumors. J. Clin. Invest. 109, 409–417 (2002).
Su, Z. et al. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J. Immunol. 174, 3798–3807 (2005).
Boczkowski, D., Nair, S. K., Nam, J. H., Lyerly, H. K. & Gilboa, E. Induction of tumor immunity and cytotoxic T lymphocyte responses using dendritic cells transfected with messenger RNA amplified from tumor cells. Cancer Res. 60, 1028–1034 (2000).
Rains, N., Cannan, R. J., Chen, W. & Stubbs, R. S. Development of a dendritic cell (DC)-based vaccine for patients with advanced colorectal cancer. Hepatogastroenterology 48, 347–351 (2001).
Caruso, D. A. et al. Results of a phase 1 study utilizing monocyte-derived dendritic cells pulsed with tumor RNA in children and young adults with brain cancer. Neuro Oncol. 6, 236–246 (2004).
Wilgenhof, S. et al. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann. Oncol. 24, 2686–2693 (2013).
Van Lint, S. et al. Optimized dendritic cell-based immunotherapy for melanoma: the TriMix-formula. Cancer Immunol. Immunother. 63, 959–967 (2014).
Van Lint, S. et al. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res. 72, 1661–1671 (2012).
Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010).
Amin, A. et al. Survival with AGS-003, an autologous dendritic cell-based immunotherapy, in combination with sunitinib in unfavorable risk patients with advanced renal cell carcinoma (RCC): phase 2 study results. J. Immunother. Cancer 3, 14 (2015).
Figlin, R. et al. Interim analysis of the phase 3 ADAPT trial evaluating rocapuldencel-T (AGS-003), an individualized immunotherapy for the treatment of newly-diagnosed patients with metastatic renal cell carcinoma (mRCC). Ann. Oncol. https://doi.org/10.1093/annonc/mdx376.003 (2017).
Fotin-Mleczek, M. et al. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 34, 1–15 (2011).
Alberer, M. et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet 390, 1511–1520 (2017).
Kubler, H. et al. Self-adjuvanted mRNA vaccination in advanced prostate cancer patients: a first-in-man phase I/IIa study. J. Immunother. Cancer 3, 26 (2015).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT01817738 (2013).
Kreiter, S. et al. Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res. 70, 9031–9040 (2010).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02035956 (2014).
Diken, M. et al. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 18, 702–708 (2011).
Tureci, O. et al. Targeting the heterogeneity of cancer with individualized neoepitope vaccines. Clin. Cancer Res. 22, 1885–1896 (2016).
Carreno, B. M. et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808 (2015).
Zhang, X., Sharma, P. K., Peter Goedegebuure, S. & Gillanders, W. E. Personalized cancer vaccines: targeting the cancer mutanome. Vaccine 35, 1094–1100 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02510950 (2015).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02950766 (2016).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02287428 (2014).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02600949 (2015).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02721043 (2016).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02419170 (2015).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT00683670 (2008).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT01970358 (2013).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02427581 (2015).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02897765(2016).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03359239 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03166254 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03068832 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03223103 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03219450 (2017).
Kenter, G. G. et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia. N. Engl. J. Med. 361, 1838–1847 (2009).
Kranz, L. M. et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). This article describes an optimal liposome–RNA complex formulation for systemic delivery of RNA to DCs in vivo. The RNA encodes specific tumour neoantigens and can also be recognized by PRRs in DCs and macrophages, favouring the induction of a potent antitumour immune response.
Kreiter, S. et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696 (2015).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017). This is the first report on the first-in-class personalized RNA-encoded poly-neoantigen vaccine in patients with melanoma, with a workflow of point mutation identification, neoantigen computational prediction and design and production of a patient-specific RNA-based vaccine.
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03313778 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03289962 (2017).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Ruckman, J. et al. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 273, 20556–20567 (1998).
Rohloff, J. C. et al. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids 3, e201 (2014).
Klussmann, S., Nolte, A., Bald, R., Erdmann, V. A. & Furste, J. P. Mirror-image RNA that binds D-adenosine. Nat. Biotechnol. 14, 1112–1115 (1996).
Gao, S., Zheng, X., Jiao, B. & Wang, L. Post-SELEX optimization of aptamers. Anal. Bioanal Chem. 408, 4567–4573 (2016).
Cho, M. et al. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl Acad. Sci. USA 107, 15373–15378 (2010).
Alam, K. K., Chang, J. L. & Burke, D. H. FASTAptamer: a bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol. Ther. Nucleic Acids 4, e230 (2015).
Santulli-Marotto, S., Nair, S. K., Rusconi, C., Sullenger, B. & Gilboa, E. Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 63, 7483–7489 (2003).
Berezhnoy, A. et al. Isolation and optimization of murine IL-10 receptor blocking oligonucleotide aptamers using high-throughput sequencing. Mol. Ther. 20, 1242–1250 (2012).
Hervas-Stubbs, S. et al. Identification of TIM3 2′-fluoro oligonucleotide aptamer by HT-SELEX for cancer immunotherapy. Oncotarget 7, 4522–4530 (2016).
Gefen, T. et al. A TIM-3 oligonucleotide aptamer enhances t cell functions and potentiates tumor immunity in mice. Mol. Ther. 25, 2280–2288 (2017).
Soldevilla, M. M. et al. Identification of LAG3 high affinity aptamers by HT-SELEX and Conserved Motif Accumulation (CMA). PLOS One 12, e0185169 (2017).
Ajona, D. et al. A combined PD-1/C5a blockade synergistically protects against lung cancer growth and metastasis. Cancer Discov. 7, 694–703 (2017).
McNamara, J. O. et al. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Invest. 118, 376–386 (2008). This article describes the first agonistic aptamer generated by multimerization and/or dimerization of a 4-1BB binding aptamer.
Dollins, C. M. et al. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 15, 675–682 (2008).
Pastor, F. et al. CD28 aptamers as powerful immune response modulators. Mol. Ther. Nucleic Acids 2, e98 (2013).
Soldevilla, M. M. et al. 2-Fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia. Biomaterials 67, 274–285 (2015).
Lee, S. W., Salek-Ardakani, S., Mittler, R. S. & Croft, M. Hypercostimulation through 4-1BB distorts homeostasis of immune cells. J. Immunol. 182, 6753–6762 (2009).
Pastor, F., Kolonias, D., McNamara, J. O. 2nd & Gilboa, E. Targeting 4-1BB costimulation to disseminated tumor lesions with bi-specific oligonucleotide aptamers. Mol. Ther. 19, 1878–1886 (2011).
Schrand, B. et al. Targeting 4-1BB costimulation to the tumor stroma with bispecific aptamer conjugates enhances the therapeutic index of tumor immunotherapy. Cancer Immunol. Res. 2, 867–877 (2014).
Schrand, B. et al. Radiation-induced enhancement of antitumor T cell immunity by VEGF-targeted 4-1BB costimulation. Cancer Res. 77, 1310–1321 (2017). This article describes the application of bispecific VEGF–4-1BB aptamer to target 4-1BB co-stimulation to the tumour stroma after VEGF release by local radiation.
Soldevilla, M. M. et al. MRP1-CD28 bi-specific oligonucleotide aptamers: target costimulation to drug-resistant melanoma cancer stem cells. Oncotarget 7, 23182–23196 (2016).
McNamara, J. O. 2nd et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 24, 1005–1015 (2006).
Dassie, J. P. et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 27, 839–849 (2009).
Pastor, F., Kolonias, D., Giangrande, P. H. & Gilboa, E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 465, 227–230 (2010). This article describes a therapeutic approach to increase tumour antigenicity by inhibiting NMD by using an aptamer–siRNA conjugate.
Lykke-Andersen, S. & Jensen, T. H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677 (2015).
Melero, I., Murillo, O., Dubrot, J., Hervas-Stubbs, S. & Perez-Gracia, J. L. Multi-layered action mechanisms of CD137 (4-1BB)-targeted immunotherapies. Trends Pharmacol. Sci. 29, 383–390 (2008).
Berezhnoy, A., Castro, I., Levay, A., Malek, T. R. & Gilboa, E. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J. Clin. Invest. 124, 188–197 (2014).
Rajagopalan, A., Berezhnoy, A., Schrand, B., Puplampu-Dove, Y. & Gilboa, E. Aptamer-targeted attenuation of IL-2 signaling in CD8(+) T cells enhances antitumor immunity. Mol. Ther. 25, 54–61 (2017).
Herrmann, A. et al. CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. J. Clin. Invest. 124, 2977–2987 (2014).
Lozano, T. et al. Targeting inhibition of Foxp3 by a CD28 2′-Fluro oligonucleotide aptamer conjugated to P60-peptide enhances active cancer immunotherapy. Biomaterials 91, 73–80 (2016).
Zhou, J. & Rossi, J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT00950638 (2009).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT01940900 (2013).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03168139 (2017).
Zboralski, D., Hoehlig, K., Eulberg, D., Fromming, A. & Vater, A. Increasing tumor-infiltrating T Cells through inhibition of CXCL12 with NOX-A12 synergizes with PD-1 blockade. Cancer Immunol. Res. 5, 950–956 (2017).
Chester, C., Ambulkar, S. & Kohrt, H. E. 4-1BB agonism: adding the accelerator to cancer immunotherapy. Cancer Immunol. Immunother. 65, 1243–1248 (2016).
Suntharalingam, G. et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 355, 1018–1028 (2006).
Grupp, S. A. et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 368, 1509–1518 (2013).
Locke, F. L. et al. Phase 1 results of ZUMA-1: a multicenter study of KTE-C19 anti-CD19 CAR T cell therapy in refractory aggressive lymphoma. Mol. Ther. 25, 285–295 (2017).
Qasim, W. et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 9, eaaj2013 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02735083 (2016).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT03166878 (2017).
Boissel, L. et al. Comparison of mRNA and lentiviral based transfection of natural killer cells with chimeric antigen receptors recognizing lymphoid antigens. Leuk. Lymphoma 53, 958–965 (2012).
Lee, J. M. et al. Direct and indirect antitumor effects by human peripheral blood lymphocytes expressing both chimeric immune receptor and interleukin-2 in ovarian cancer xenograft model. Cancer Gene Ther. 17, 742–750 (2010).
Stadler, C. R. et al. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 23, 815–817 (2017). This article presents the proof of concept of the feasibility of using bispecific antibodies encoded by an RNA therapeutic molecule. This approach sustains high levels of endogenous recombinant bispecific antibody production, eliciting a robust antitumour response.
Ghafouri-Fard, S. siRNA and cancer immunotherapy. Immunotherapy 4, 907–917 (2012).
Bachmaier, K. et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature 403, 211–216 (2000).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT02166255 (2014).
Qian, Y. et al. Molecular-targeted immunotherapeutic strategy for melanoma via dual-targeting nanoparticles delivering small interfering RNA to tumor-associated macrophages. ACS Nano 11, 9536–9549 (2017).
Yin, H. et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 35, 1179–1187 (2017).
Cubillos-Ruiz, J. R. et al. Reprogramming tumor-associated dendritic cells in vivo using miRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 72, 1683–1693 (2012).
Jain, R. et al. MicroRNAs enable mRNA therapeutics to selectively program cancer cells to self-destruct. Nucleic Acid Ther. https://doi.org/10.1089/nat.2018.0734 (2018).
Keene, J. D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 (2007).
Zubiaga, A. M., Belasco, J. G. & Greenberg, M. E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 15, 2219–2230 (1995).
Sadler, A. J. & Williams, B. R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 (2008).
Allerson, C. R. et al. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 48, 901–904 (2005).
Harborth, J. et al. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid. Drug Dev. 13, 83–105 (2003).
Strenkowska, M. et al. Cap analogs modified with 1,2-dithiodiphosphate moiety protect mRNA from decapping and enhance its translational potential. Nucleic Acids Res. 44, 9578–9590 (2016).
Kowalska, J. et al. Synthesis, properties, and biological activity of boranophosphate analogs of the mRNA cap: versatile tools for manipulation of therapeutically relevant cap-dependent processes. Nucleic Acids Res. 42, 10245–10264 (2014).
Stepinski, J., Waddell, C., Stolarski, R., Darzynkiewicz, E. & Rhoads, R. E. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA 7, 1486–1495 (2001).
Wang, Z., Day, N., Trifillis, P. & Kiledjian, M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 19, 4552–4560 (1999).
Presnyak, V. et al. Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124 (2015).
Mockey, M. et al. mRNA transfection of dendritic cells: synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem. Biophys. Res. Commun. 340, 1062–1068 (2006).
Svitkin, Y. V. et al. N1-methyl-pseudouridine in mRNA enhances translation through eIF2alpha-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 45, 6023–6036 (2017).
US National Library of Medicine. ClinicalTrials.gov. https://www.clinicalTrials.gov/show/NCT00204607 (2005).
Heyes, J., Palmer, L., Bremner, K. & MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control Release 107, 276–287 (2005).
Coelho, T. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 369, 819–829 (2013).
Jayaraman, M. et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed Engl. 51, 8529–8533 (2012). This manuscript identifies DLin-MC3-DMA as one of the most potent ionizable amino lipids to deliver RNA-based drugs to the liver.
Zimmermann, T. S. et al. Clinical proof of concept for a novel hepatocyte-targeting GalNAc-siRNA conjugate. Mol. Ther. 25, 71–78 (2017).
Liang, F. et al. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 25, 2635–2647 (2017).
Ramishetti, S. et al. Systemic gene silencing in primary T lymphocytes using targeted lipid nanoparticles. ACS Nano 9, 6706–6716 (2015).
Wengerter, B. C. et al. Aptamer-targeted antigen delivery. Mol. Ther. 22, 1375–1387 (2014).
Ishida, T. et al. Injection of PEGylated liposomes in rats elicits PEG-specific IgM, which is responsible for rapid elimination of a second dose of PEGylated liposomes. J. Control Release 112, 15–25 (2006).
Naito, Y., Yoshimura, J., Morishita, S. & Ui-Tei, K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics 10, 392 (2009).
Hannus, M. et al. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 42, 8049–8061 (2014).
Jackson, A. L. et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12, 1197–1205 (2006).
Carthew, R. W. & Sontheimer, E. J. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009).
Han, J. et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125, 887–901 (2006).
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004).
Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R. & Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110, 563–574 (2002).
Fabian, M. R., Sonenberg, N. & Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 (2010).
Kim, D. H. et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23, 222–226 (2005).
Iwasaki, Y. W., Siomi, M. C. & Siomi, H. PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433 (2015).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949.e15 (2017).
Bol, K. F., Schreibelt, G., Gerritsen, W. R., de Vries, I. J. & Figdor, C. G. Dendritic cell-based immunotherapy: state of the art and beyond. Clin. Cancer Res. 22, 1897–1906 (2016).
Tacken, P. J., de Vries, I. J., Torensma, R. & Figdor, C. G. Dendritic-cell immunotherapy: from ex vivo loading to in vivo targeting. Nat. Rev. Immunol. 7, 790–802 (2007).
Kastenmuller, W., Kastenmuller, K., Kurts, C. & Seder, R. A. Dendritic cell-targeted vaccines—hope or hype? Nat. Rev. Immunol. 14, 705–711 (2014).
Lincoff, A. M. et al. Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): a randomised clinical trial. Lancet 387, 349–356 (2016).
Titze-de-Almeida, R., David, C. & Titze-de-Almeida, S. S. The race of 10 synthetic RNAi-based drugs to the pharmaceutical market. Pharm. Res. 34, 1339–1363 (2017).
Sinha, G. Regado's aptamer lines up against anticoagulants. Nat. Biotechnol. 31, 1060 (2013).
Oney, S. et al. Development of universal antidotes to control aptamer activity. Nat. Med. 15, 1224 (2009).
Acknowledgements
This work was supported by Worldwide Cancer Research grants 15–1146 and 15–1208, the Asociación Española Contra el Cáncer (AECC) Foundation under grant GCB15152947MELE, Red Temática de Investigación Cooperativa en Cáncer under grants RD12/0036/0040 and RD12/0036/0062, Fondo de Investigación Sanitaria-Fondo Europeo de Desarrollo Regional (FEDER) under grants PI14/01686, PI13/00207, PI16/00668 and PI17/00372, the H2020 PROCROP project under grant 635122, the Melanoma Research Alliance under grant 509510 and the Fundación Ramón Areces under grant CIVP18A3916. F.P. is supported by Ramón y Cajal (10699). P.B. is supported by a Miguel Servet II (CPII15/00004) contract from the Instituto de Salud Carlos III.
Author information
Authors and Affiliations
Contributions
I.M., F.P., P.B. and I.E. researched data for the article and I.M., F.P. and P.B wrote the article. All of the authors provided substantial contribution to the discussion of the content and reviewed and edited the article before submission.
Corresponding author
Ethics declarations
Competing interests
I.M. declares financial and non-financial competing interests. I.M. reports receiving commercial research funding (grants) from Bristol-Myers Squibb (BMS) and Roche and serves as a consultant and/or advisory board member for Alligator, AstraZeneca, Bioncotech, BMS, F-STAR, Genmab, Incyte, Merck Serono, Molecular Partners, Roche–Genentech and Tusk. E.G. is founder of Sebastian Biopharma that licensed IP from the University of Miami to develop methods of inducing neoantigens. U.S. is CEO and co-founder of BioNTech, a company that develops mRNA therapeutics.
Glossary
- RNA moieties
-
Defined structures in RNA that determine its binding and functional activity.
- Promoter
-
A specific DNA sequence upstream of the gene that is recognized by the RNA polymerase complex to initiate the transcription process.
- Polyadenylation
-
Incorporation of a tail of multiple adenosine monophosphates at the 3′ end of the nascent mRNA, thereby conferring stability.
- Splicing
-
A process in eukaryotes by which the nascent transcribed mRNA is edited by removing part of the RNA (introns) and keeping the protein-encoding sequence (exons).
- Alternative splicing
-
A variation in the splicing process to include or exclude different exons such that a single gene can encode for multiple translated proteins.
- Ribonucleoproteins
-
Conjugates of RNA and protein that usually have enzymatic or scaffold functions.
- Lariat RNAs
-
Circular introns released during the splicing process in the nucleus.
- Codon rewiring
-
Changes in codon reassignment during the translation process.
- Kozak sequence
-
A conserved sequence within the mRNA (ACCAUGG) that leads the ribosome to initiate the translation of the encoded protein.
- Nonsense-mediated mRNA decay
-
(NMD). An RNA surveillance mechanism that identifies and eliminates mRNA with premature stop codons.
- Toll-like receptor
-
(TLR). A type of pattern recognition receptor named because of homology to the fruitfly protein Toll. Such structures on the cell surface or in endosomes detect moieties that denote the presence of danger signals.
- RNA helicases
-
A group of enzymes that rearrange RNA folding structures usually by unwinding the double-stranded RNA helix.
- Type I interferon
-
A group of interferon proteins including interferon-β (IFNβ) and the different subtypes of IFNα, which are cytokines with antiviral, antitumour and multiple immunomodulatory activities.
- Dendritic cells
-
(DCs). Professional antigen-presenting cells of leukocyte lineage. Several subsets are specialized at inducing and sustaining different types of immune response.
- Immune-desert tumours
-
Tumours with lack of infiltration of immune cells due to defects in the priming phase of the antitumour immune response or problems in leukocyte trafficking.
- T cell receptor
-
(TCR). A transmembrane protein expressed on T lymphocytes that recognizes peptides presented in the context of major histocompatibility complex (MHC) class I and MHC class II proteins. TCR-encoding genes are clonally rearranged, constituting an antigen recognition repertoire.
- C12-200 lipidoid
-
An epoxide-derived, oligoamine-containing, lipid-like compound used to deliver small interfering RNA to the liver.
- DNA cassettes
-
DNA fragments that contain a gene and the regulatory elements required for the gene expression in the transfected cell.
- Partial tolerance
-
A status of unresponsiveness to a given specific antigen as a result of previous exposure to such antigen. Degrees of tolerance to a given antigen are possible, and tolerance relies on the dysfunction or elimination of specific T cells.
- Protamine
-
An arginine-rich nucleoprotein that condensates DNA through electrostatic interactions.
- Immune checkpoint
-
A co-inhibitory molecule that reduces the proliferation, differentiation and effector functions of lymphocytes.
- Antisense antidotes
-
Molecules that absorb nucleic acid aptamer, disrupting its structure and therefore its function. This is achieved by complementary antisense sequence205 or positively charged polymers206.
- Gene-editing nucleases
-
Engineered nucleases that modify the genome by targeting specific genomic sequences.
- Nucleofection
-
Transfection of RNA or DNA to the nucleus and cytosol by an electroporation-based approach using a Nucleofector device.
- Infusion reactions
-
Signs or symptoms that occur during the infusion of a therapeutic agent or on the first day of administration. Clinical manifestations vary in severity and can include many different symptoms involving different body systems
Rights and permissions
About this article
Cite this article
Pastor, F., Berraondo, P., Etxeberria, I. et al. An RNA toolbox for cancer immunotherapy. Nat Rev Drug Discov 17, 751–767 (2018). https://doi.org/10.1038/nrd.2018.132
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2018.132
This article is cited by
-
Neoantigen vaccine nanoformulations based on Chemically synthesized minimal mRNA (CmRNA): small molecules, big impact
npj Vaccines (2024)
-
High-throughput barcoding of nanoparticles identifies cationic, degradable lipid-like materials for mRNA delivery to the lungs in female preclinical models
Nature Communications (2024)
-
mRNA-based cancer therapeutics
Nature Reviews Cancer (2023)
-
Charge-altering releasable transporters enhance mRNA delivery in vitro and exhibit in vivo tropism
Nature Communications (2023)
-
Aptamer-based rapid diagnosis for point-of-care application
Microfluidics and Nanofluidics (2023)