Key Points
-
DNA-encoded chemical library technologies are increasingly being adopted in drug discovery for hit and lead generation. DNA-encoded chemistry enables the exploration of chemical spaces four to five orders of magnitude more deeply than is achievable by traditional high-throughput screening methods.
-
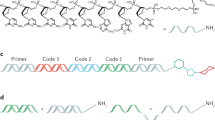
DNA-encoded chemical library technology involves the creation of large mixtures of small molecules that are encoded with sequences or single-stranded or double-stranded DNA. High-affinity hits from such mixtures are identifiable by sequencing the DNA tags associated with each compound.
-
DNA-encoded library technology began with a publication by Brenner and Lerner in 1992. The technology has subsequently evolved to be practised by several large pharmaceutical and biotechnology companies.
-
DNA-directed synthesis is a related approach. In this method, the specificity of DNA base pairing serves for both encoding and synthesis.
-
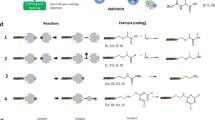
DNA-encoded library synthesis as it is currently practised is based on reactions that are tolerant to water.
-
The creation of hundreds of millions of DNA-encoded library compounds is less expensive and more feasible than assembling a library of single compounds on milligram scale.
Abstract
DNA-encoded chemical library technologies are increasingly being adopted in drug discovery for hit and lead generation. DNA-encoded chemistry enables the exploration of chemical spaces four to five orders of magnitude more deeply than is achievable by traditional high-throughput screening methods. Operation of this technology requires developing a range of capabilities including aqueous synthetic chemistry, building block acquisition, oligonucleotide conjugation, large-scale molecular biological transformations, selection methodologies, PCR, sequencing, sequence data analysis and the analysis of large chemistry spaces. This Review provides an overview of the development and applications of DNA-encoded chemistry, highlighting the challenges and future directions for the use of this technology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
30 January 2017
The details of reference 119 were incorrect and there were also some minor typographical errors. These errors have been corrected in the online version of the article.
References
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat. Rev. Drug Discov. 9, 203–214 (2010).
Shelat, A. A. & Guy, R. K. Scaffold composition and biological relevance of screening libraries. Nat. Chem. Biol. 3, 442–446 (2007).
Reymond, J. L. The Chemical Space Project. Acc. Chem. Res. 48, 722–730 (2015).
Furka, A., Sebestyen, F., Asgedom, M. & Dibo, G. in Trends in Medicinal Chemistry '88: Proceedings of the Xth International Symposium on Medicinal Chemistry, Budapest, 15–19 August 1988 Vol. 9 (eds van der Goot, H. et al.) 288 (Elsevier, 1989).
Czarnik, A. W. Encoding methods for combinatorial chemistry. Curr. Opin. Chem. Biol. 1, 60–66 (1997).
Geysen, H. M. et al. Isotope or mass encoding of combinatorial libraries. Chem. Biol. 3, 679–688 (1996).
Ohlmeyer, M. H. J. et al. Complex synthetic chemical libraries indexed with molecular tags. Proc. Natl Acad. Sci. USA 90, 10922–10926 (1993).
Nicolaou, K. C., Xiao, X. Y., Parandoosh, Z., Senyei, A. & Nova, M. P. Radiofrequency encoded combinatorial chemistry. Angew. Chem. Int. Ed. 34, 2289–2291 (1995).
Geysen, H. M., Meloen, R. H. & Barteling, S. J. Use of peptide-synthesis to probe viral-antigens for epitopes to a resolution of a single amino-acid. Proc. Natl Acad. Sci. USA 81, 3998–4002 (1984).
Baca, M., Presta, L. G., O'Connor, S. J. & Wells, J. A. Antibody humanization using monovalent phage display. J. Biol. Chem. 272, 10678–10684 (1997).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Hanes, J. & Pluckthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl Acad. Sci. USA 94, 4937–4942 (1997).
Hoogenboom, H. R. & Winter, G. By-passing immunisation. Human antibodies from synthetic repertoires of germline VH gene segments rearranged in vitro. J. Mol. Biol. 227, 381–388 (1992).
Roberts, R. W. & Szostak, J. W. RNA–peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl Acad. Sci. USA 94, 12297–12302 (1997).
Smith, G. P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Brenner, S. & Lerner, R. A. Encoded combinatorial chemistry. Proc. Natl Acad. Sci. USA 89, 5381–5383 (1992). Describes the concept of a DEL for the first time, including how such a library can be interrogated using affinity-mediated selection and how enriched compounds can be amplified and ultimately identified by sequencing.
Needels, M. C. et al. Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proc. Natl Acad. Sci. USA 90, 10700–10704 (1993). Describes the first use of a DEL to discover functional library members, in this case known antibody epitopes from an encoded peptide library.
Schroeder, G. K., Lad, C., Wyman, P., Williams, N. H. & Wolfenden, R. The time required for water attack at the phosphorus atom of simple phosphodiesters and of DNA. Proc. Natl Acad. Sci. USA 103, 4052–4055 (2006).
Clark, M. A. Selecting chemicals: the emerging utility of DNA-encoded libraries. Curr. Opin. Chem. Biol. 14, 396–403 (2010).
Buller, F. et al. Design and synthesis of a novel DNA-encoded chemical library using Diels-Alder cycloadditions. Bioorg. Med. Chem. Lett. 18, 5926–5931 (2008). The first report of using a DNA-recorded split-and-pool library for ligand discovery.
Dumelin, C. E. et al. A portable albumin binder from a DNA-encoded chemical library. Angew. Chem. Int. Ed. 47, 3196–3201 (2008).
Bentley, D. R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Ciobanu, M. et al. Selection of a synthetic glycan oligomer from a library of DNA-templated fragments against DC-SIGN and inhibition of HIV gp120 binding to dendritic cells. Chem. Commun. 47, 9321–9323 (2011).
Debaene, F., Da Silva, J. A., Pianowski, Z., Duran, F. J. & Winssinger, N. Expanding the scope of PNA-encoded libraries: divergent synthesis of libraries targeting cysteine, serine and metalloproteases as well as tyrosine phosphatases. Tetrahedron 63, 6577–6586 (2007).
Svensen, N., Diaz-Mochon, J. J. & Bradley, M. Decoding a PNA encoded peptide library by PCR: the discovery of new cell surface receptor ligands. Chem. Biol. 18, 1284–1289 (2011).
Svensen, N., Diaz-Mochon, J. J. & Bradley, M. Encoded peptide libraries and the discovery of new cell binding ligands. Chem. Commun. 47, 7638–7640 (2011).
Annis, D. A. et al. An affinity selection–mass spectrometry method for the identification of small molecule ligands from self-encoded combinatorial libraries — discovery of a novel antagonist of E-coli dihydrofolate reductase. Int. J. Mass Spectrom. 238, 77–83 (2004).
Annis, D. A. et al. Method for quantitative protein–ligand affinity measurements in compound mixtures. Anal. Chem. 79, 4538–4542 (2007).
Brudno, Y. & Liu, D. R. Recent progress toward the templated synthesis and directed evolution of sequence-defined synthetic polymers. Chem. Biol. 16, 265–276 (2009).
Kinoshita, Y. & Nishigaki, K. Enzymatic synthesis of code regions for encoded combinatorial chemistry (ECC). Nucleic Acids Symp. Ser. 34, 201–202 (1995). Describes the concept of the enzymatic ligation of oligonucleotide tags as a methodology for the generation of encoded chemical libraries for the first time.
Litovchick, A. et al. Encoded library synthesis using chemical ligation and the discovery of sEH inhibitors from a 334-million member library. Sci. Rep. 5, 10916 (2015).
Clark, M. A. et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 5, 647–654 (2009). One of the earliest and most complete descriptions of the synthesis and use of a DNA-recorded chemical library to find inhibitors of multiple therapeutic targets.
Deng, H. et al. Discovery, SAR, and X-ray binding mode study of BCATm inhibitors from a novel DNA-encoded library. ACS Med. Chem. Lett. 6, 919–924 (2015).
Mannocci, L. et al. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc. Natl Acad. Sci. USA 105, 17670–17675 (2008). The earliest description of deep sequencing of DNA-encoded chemical libraries.
Deng, H. et al. Discovery of highly potent and selective small molecule ADAMTS-5 inhibitors that inhibit human cartilage degradation via encoded library technology (ELT). J. Med. Chem. 55, 7061–7079 (2012).
Arico-Muendel, C. et al. Encoded library technology screening of hepatitis c virus NS4B yields a small-molecule compound series with in vitro replicon activity. Antimicrob. Agents Chemother. 59, 3450–3459 (2015).
Samain, F. et al. Tankyrase 1 inhibitors with drug-like properties identified by screening a DNA-encoded chemical library. J. Med. Chem. 58, 5143–5149 (2015).
Gartner, Z. J. et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science 305, 1601–1605 (2004). Earliest description of the use of a DNA-directed chemical library of any kind to find known inhibitors.
Hansen, M. H. et al. A yoctoliter-scale DNA reactor for small-molecule evolution. J. Am. Chem. Soc. 131, 1322–1327 (2009).
Tse, B. N., Snyder, T. M., Shen, Y. H. & Liu, D. R. Translation of DNA into a library of 13 000 synthetic small-molecule macrocycles suitable for in vitro selection. J. Am. Chem. Soc. 130, 15611–15626 (2008).
Catalano, M. J., Price, N. E. & Gates, K. S. Effective molarity in a nucleic acid-controlled reaction. Bioorg. Med. Chem. Lett. 26, 2627–2630 (2016).
Gartner, Z. J. & Liu, D. R. The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J. Am. Chem. Soc. 123, 6961–6963 (2001).
Seigal, B. A. et al. The discovery of macrocyclic XIAP antagonists from a DNA-programmed chemistry library, and their optimization to give lead compounds with in vivo antitumor activity. J. Med. Chem. 58, 2855–2861 (2015).
Kleiner, R. E., Dumelin, C. E., Tiu, G. C., Sakurai, K. & Liu, D. R. In vitro selection of a DNA-templated small-molecule library reveals a class of macrocyclic kinase inhibitors. J. Am. Chem. Soc. 132, 11779–11791 (2010). The first description of using a DTS library to find novel inhibitors of therapeutic targets.
Maianti, J. P. et al. Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature 511, 94–98 (2014).
Cao, C. et al. A DNA-templated synthesis of encoded small molecules by DNA self-assembly. xChem. Commun. 50, 10997–10999 (2014).
Petersen, L. K. et al. Novel p38α MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library. MedChemComm 7, 1332–1339 (2016). The first description of using both the yoctoReactor and the binder trap enrichment technology to find novel inhibitors of a therapeutic target.
Halpin, D. R. & Harbury, P. B. DNA display I. Sequence-encoded routing of DNA populations. PLoS Biol. 2, E173 (2004).
Halpin, D. R. & Harbury, P. B. DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution. PLoS Biol. 2, E174 (2004).
Halpin, D. R., Lee, J. A., Wrenn, S. J. & Harbury, P. B. DNA display III. Solid-phase organic synthesis on unprotected DNA. PLoS Biol. 2, E175 (2004). The first description of using DNA routing to rediscover a known ligand.
Wrenn, S. J., Weisinger, R. M., Halpin, D. R. & Harbury, P. B. Synthetic ligands discovered by in vitro selection. J. Am. Chem. Soc. 129, 13137–13143 (2007). The first description of using a DNA-directed chemical library synthesized using hybridization-mediated routing to find binders to a protein target.
Weisinger, R. M., Wrenn, S. J. & Harbury, P. B. Highly parallel translation of DNA sequences into small molecules. PLoS ONE 7, e28056 (2012).
Melkko, S., Scheuermann, J., Dumelin, C. E. & Neri, D. Encoded self-assembling chemical libraries. Nat. Biotechnol. 22, 568–574 (2004). The first description of encoded self-assembling chemical libraries for ligand discovery.
Melkko, S., Zhang, Y., Dumelin, C. E., Scheuermann, J. & Neri, D. Isolation of high-affinity trypsin inhibitors from a DNA-encoded chemical library. Angew. Chem. Int. Ed. 46, 4671–4674 (2007).
Wichert, M. et al. Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation. Nat. Chem. 7, 241–249 (2015).
Li, G. et al. Design, preparation, and selection of DNA-encoded dynamic libraries. Chem. Sci. 6, 7097–7104 (2015).
Reddavide, F. V., Lin, W. L., Lehnert, S. & Zhang, Y. X. DNA-encoded dynamic combinatorial chemical libraries. Angew. Chem. Int. Ed. 54, 7924–7928 (2015).
Daguer, J. P. et al. DNA display of fragment pairs as a tool for the discovery of novel biologically active small molecules. Chem. Sci. 6, 739–744 (2015).
Barluenga, S. et al. Novel PTP1B inhibitors identified by DNA display of fragment pairs. Bioorg. Med. Chem. Lett. 26, 1080–1085 (2016).
Scheuermann, J. et al. DNA-encoded chemical libraries for the discovery of MMP-3 inhibitors. Bioconjug. Chem. 19, 778–785 (2008).
Premstaller, A., Oberacher, H. & Huber, C. G. High-performance liquid chromatography-electrospray ionization mass spectrometry of single- and double-stranded nucleic acids using monolithic capillary columns. Anal. Chem. 72, 4386–4393 (2000).
Satz, A. L. et al. DNA compatible multistep synthesis and applications to DNA encoded libraries. Bioconjug. Chem. 26, 1623–1632 (2015).
Tian, X. et al. Development and design of the tertiary amino effect reaction for DNA-encoded library synthesis. MedChemComm 7, 1316–1322 (2016).
Li, C. J. Organic reactions in aqueous media with a focus on carbon-carbon bond formations: a decade update. Chem. Rev. 105, 3095–3165 (2005).
Weisinger, R. M., Marinelli, R. J., Wrenn, S. J. & Harbury, P. B. Mesofluidic devices for DNA-programmed combinatorial chemistry. PLoS ONE 7, e32299 (2012).
Rozenman, M. M. & Liu, D. R. DNA-templated synthesis in organic solvents. Chembiochem 7, 253–256 (2006).
Liu, K. et al. Nucleic acid chemistry in the organic phase: from functionalized oligonucleotides to DNA side chain polymers. J. Am. Chem. Soc. 136, 14255–14262 (2014).
Kalliokoski, T. Price-focused analysis of commercially available building blocks for combinatorial library synthesis. ACS Comb. Sci. 17, 600–607 (2015).
Satz, A. L. in A Handbook For DNA-Encoded Chemistry: Theory And Applications For Exploring Chemical Space (ed. Goodnow, R. A. J.) 99–121 (Wiley & Sons, 2014).
Creaser, S. P. in A Handbook For DNA-Encoded Chemistry: Theory And Applications For Exploring Chemical Space (ed. Goodnow, R. A. J.) 123–151 (Wiley & Sons, 2014).
Gilar, M. et al. Ion-pair reversed-phase high-performance liquid chromatography analysis of oligonucleotides: retention prediction. J. Chromatogr. A 958, 167–182 (2002).
Potier, N., Vandorsselaer, A., Cordier, Y., Roch, O. & Bischoff, R. Negative electrospray-ionization mass-spectrometry of synthetic and chemically-modified oligonucleotides. Nucleic Acids Res. 22, 3895–3903 (1994).
Hargiss, L. O. et al. Reaction profiling by ultra high-pressure liquid chromatography/time-of-flight mass spectrometry in support of the synthesis of DNA-encoded libraries. J. Chromatogr. B 971, 120–125 (2014).
Feinberg, J. A. in A Handbook For DNA-Encoded Chemistry: Theory And Applications For Exploring Chemical Space (ed. Goodnow, R. A. J.) 201–212 (Wiley & Sons, 2014).
Decurtins, W. et al. Automated screening for small organic ligands using DNA-encoded chemical libraries. Nat. Protoc. 11, 764–780 (2016).
Blakskjaer, P., Heitner, T. & Hansen, N. J. V. Fidelity by design: yoctoReactor and binder trap enrichment for small-molecule DNA-encoded libraries and drug discovery. Curr. Opin. Chem. Biol. 26, 62–71 (2015).
Denton, K. E. & Krusemark, C. J. Crosslinking of DNA-linked ligands to target proteins for enrichment from DNA-encoded libraries. MedChemComm 7, 2020–2027 (2016).
Zhao, P. et al. Selection of DNA-encoded small molecule libraries against unmodified and non-immobilized protein targets. Angew. Chem. Int. Ed. 53, 10056–10059 (2014).
McGregor, L. M., Jain, T. & Liu, D. R. Identification of ligand–target pairs from combined libraries of small molecules and unpurified protein targets in cell lysates. J. Am. Chem. Soc. 136, 3264–3270 (2014).
McGregor, L. M., Gorin, D. J., Dumelin, C. E. & Liu, D. R. Interaction-dependent PCR: identification of ligand-target pairs from libraries of ligands and libraries of targets in a single solution-phase experiment. J. Am. Chem. Soc. 132, 15522–15224 (2010).
Bao, J. Y. et al. Prediction of protein–DNA complex mobility in gel-free capillary electrophoresis. Anal. Chem. 87, 2474–2479 (2015).
Wu, Z. N. et al. Cell-based selection expands the utility of DNA-encoded small-molecule library technology to cell surface drug targets: identification of novel antagonists of the NK3 tachykinin receptor. ACS Comb. Sci. 17, 722–731 (2015). The first example of using a DNA-encoded chemical library to find inhibitors of a cell surface target; the inhibitors were discovered using the target in a cell surface environment.
Goodnow, R.A. Jr & Davie, C. P. 2015 First Boston Symposium of Encoded Library Platforms. MedChemComm 7, 1268–1270 (2016).
Zambaldo, C., Daguer, J. P., Saarbach, J., Barluenga, S. & Winssinger, N. Screening for covalent inhibitors using DNA-display of small molecule libraries functionalized with cysteine reactive moieties. MedChemComm 7, 1340–1351 (2016).
Harris, P. A. et al. DNA-encoded library screening identifies benzo[b][1,4]oxazepin-4-ones as highly potent and monoselective receptor interacting protein 1 kinase inhibitors. J. Med. Chem. 59, 2163–2178 (2016).
Kollmann, C. S. et al. Application of encoded library technology (ELT) to a protein–protein interaction target: discovery of a potent class of integrin lymphocyte function-associated antigen 1 (LFA-1) antagonists. Bioorg. Med. Chem. 22, 2353–2365 (2014).
Phelps, C. in Drug Discovery Chemistry April 19–22, 2016. San Diego, CA (Cambridge Innovation Institute, 2016).
Krusemark, C. J., Tilmans, N. P., Brown, P. O. & Harbury, P. B. Directed chemical evolution with an outsized genetic code. PLoS ONE 11, e0154765 (2016).
Charnley, A. K. et al. Crystal structures of human RIP2 kinase catalytic domain complexed with ATP-competitive inhibitors: foundations for understanding inhibitor selectivity. Bioorg. Med. Chem. 23, 7000–7006 (2015).
Deng, H. F. et al. Discovery and optimization of potent, selective, and in vivo efficacious 2-aryl benzimidazole BCATm inhibitors. ACS Med. Chem. Lett. 7, 379–384 (2016).
Ding, Y. et al. Discovery of potent and selective inhibitors for ADAMTS-4 through DNA-encoded library technology (ELT). ACS Med. Chem. Lett. 6, 888–893 (2015).
Lewis, H. D. et al. Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat. Chem. Biol. 11, 189–191 (2015).
Mandal, P. et al. RIP3 induces apoptosis independent of pronecrotic kinase activity. Mol. Cell 56, 481–495 (2014).
Wood, E. R. et al. The role of phosphodiesterase 12 (PDE12) as a negative regulator of the innate immune response and the discovery of antiviral inhibitors. J. Biol. Chem. 290, 19681–19696 (2015).
Yang, H. et al. Discovery of a potent class of PI3Kα inhibitors with unique binding mode via encoded library technology (ELT). ACS Med. Chem. Lett. 6, 531–536 (2015).
Franzini, R. M., Nauer, A., Scheuermann, J. & Neri, D. Interrogating target-specificity by parallel screening of a DNA-encoded chemical library against closely related proteins. Chem. Commun. 51, 8014–8016 (2015).
Satz, A. L. DNA encoded library selections and insights provided by computational simulations. ACS Chem. Biol. 10, 2237–2245 (2015).
Encinas, L. et al. Encoded library technology as a source of hits for the discovery and lead optimization of a potent and selective class of bactericidal direct inhibitors of Mycobacterium tuberculosis InhA. J. Med. Chem. 57, 1276–1288 (2014).
Buller, F. et al. Selection of carbonic anhydrase IX inhibitors from one million DNA-encoded compounds. ACS Chem. Biol. 6, 336–344 (2011).
Gilmartin, A. G. et al. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nat. Chem. Biol. 10, 181–187 (2014).
Disch, J. S. et al. Discovery of thieno[3,2-d]pyrimidine-6-carboxamides as potent inhibitors of SIRT1, SIRT2, and SIRT3. J. Med. Chem. 56, 3666–3679 (2013).
Franzini, R. M. et al. Identification of structure–activity relationships from screening a structurally compact DNA-encoded chemical library. Angew. Chem. Int. Ed. 54, 3927–3931 (2015).
Eidam, O. & Satz, A. L. Analysis of the productivity of DNA encoded libraries. MedChemComm 7, 1323–1331 (2016).
Gaulton, A. et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 (2012).
Papadatos, G. et al. SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res. 44, D1220–D1228 (2016).
Goodnow, R. A. J. in A Handbook for DNA-Encoded Chemistry: Theory and Applications for Exploring Chemical Space (ed. Goodnow, R. A. J.) 417–426 (Wiley & Sons, 2014).
Kanan, M. W., Rozenman, M. M., Sakurai, K., Snyder, T. M. & Liu, D. R. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature 431, 545–549 (2004).
Brown, D. G. & Bostrom, J. Analysis of past and present synthetic methodologies on medicinal chemistry: where have all the new reactions gone? J. Med. Chem. 59, 4443–4458 (2016).
Franzini, R. M. & Randolph, C. Chemical space of DNA-encoded libraries. J. Med. Chem. 59, 6629–6644 (2016).
Li, Y. Z., Zhao, P., Zhang, M. D., Zhao, X. Y. & Li, X. Y. Multistep DNA-templated synthesis using a universal template. J. Am. Chem. Soc. 135, 17727–17730 (2013).
He, Y. & Liu, D. R. A sequential strand-displacement strategy enables efficient six-step DNA-templated synthesis. J. Am. Chem. Soc. 133, 9972–9975 (2011).
He, Y. & Liu, D. R. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nat. Nanotechnol. 5, 778–782 (2010).
Meng, W. et al. An autonomous molecular assembler for programmable chemical synthesis. Nat. Chem. 8, 542–548 (2016).
Arico-Muendel, C. C. From haystack to needle: finding value with DNA encoded library technology at GSK. MedChemComm 7, 1898–1909 (2016).
Ottl, J. in A Handbook for DNA-Encoded Chemistry: Theory and Applications for Exploring Chemical Space (ed. Goodnow, R. A. J.) 319–345 (Wiley & Sons, 2014).
Concha, N. et al. Discovery and characterization of a class of pyrazole inhibitors of bacterial undecaprenyl pyrophosphate synthase. J. Med. Chem. 59, 7299–7304 (2016).
Soutter, H. H. et al. Discovery of novel, co-factor specific, bactericidal Mycobacterium tuberculosis InhA inhibitors using DNA-encoded library technology. Proc. Natl Acad. Sci. USA 113, E7880–E7889 (2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
At the time of writing this manuscript R.A.G. was an Executive Director at AstraZeneca. C.E.D. is an investigator at Novartis Pharma, and A.D.K. is a Director at X-Chem.
Related links
DATABASES
Glossary
- Lead
-
A hit compound that has been characterized in terms of its drug-like properties and is likely to be a therapeutically useful starting point for improvements in potency, selectivity and pharmacokinetic profile.
- High-throughput screening
-
(HTS). An assay of many compounds in parallel, often by automated means.
- Hits
-
Compounds identified in a primary screen that upon off-DNA resynthesis show reproducible biochemical activity, which is confirmed in an orthogonal assay.
- Split-and-pool synthesis
-
A combinatorial chemistry practice for creating large numbers of compounds. After the separate reaction of a first set of reagents with a common chemical transformation, the products are pooled together, resulting in a mixture of the products. This mixture is then re-arrayed for another round of separate synthesis, followed by pooling, resulting in the combination of all possible products of the two sets of building blocks.
- DNA-encoded chemistry
-
(DEC). Encoded chemistry with encoded information stored in DNA sequences.
- DNA-encoded library
-
(DEL). A collection of variants with distinguishing characteristics encoded in DNA.
- Affinity-mediated selection
-
Enrichment for binding to a target, usually via target immobilization.
- PCR amplification
-
The use of a template-dependent thermostable polymerase enzyme to amplify DNA through recursive cycles of primer binding, primer extension and thermal denaturation.
- Off-DNA resynthesis
-
The resynthesis of compounds identified in a primary screen for the purposes of assessing their biochemical or biophysical activity in the absence of DNA.
- DNA encoding
-
The establishment of a defined relationship between DNA sequence and chemical history.
- Building blocks
-
Chemical reagents containing at least one reactive handle.
- DNA tags
-
Specific sequences of DNA used to encode a specified chemical step.
- Encoded self-assembled chemistry
-
Molecular fragments that are held in co-proximity via the hybridization of complementary DNA.
- DEAE sepharose
-
Sepharose beads covalently linked to diethylaminoethyl groups for the reversible capture of DNA.
- Disynthon
-
A compound library member resulting from the combination of two types of building block.
- Rule of 5
-
A compound is deemed compliant to the Lipinski 'rule of 5' if its molecular weight is not greater than 500 Da, if its logP is not greater than 5 and if its counts of hydrogen bond donors and acceptors are not greater than 5 and 10, respectively
- Fragment-based lead discovery
-
Discovery of near leads through the assembly of small fragments that bind in a highly efficient manner.
Rights and permissions
About this article
Cite this article
Goodnow, R., Dumelin, C. & Keefe, A. DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat Rev Drug Discov 16, 131–147 (2017). https://doi.org/10.1038/nrd.2016.213
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2016.213
This article is cited by
-
Protein-templated ligand discovery via the selection of DNA-encoded dynamic libraries
Nature Chemistry (2024)
-
Trio-pharmacophore DNA-encoded chemical library for simultaneous selection of fragments and linkers
Nature Communications (2023)
-
A DNA-encoded chemical library based on chiral 4-amino-proline enables stereospecific isozyme-selective protein recognition
Nature Chemistry (2023)
-
Protein degraders enter the clinic — a new approach to cancer therapy
Nature Reviews Clinical Oncology (2023)
-
Control of the antitumour activity and specificity of CAR T cells via organic adapters covalently tethering the CAR to tumour cells
Nature Biomedical Engineering (2023)