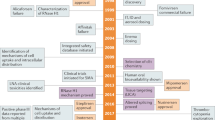
Key Points
-
Nucleic acid aptamers, often termed chemical antibodies, are short, single-stranded DNA or RNA molecules (20–100 nucleotides in length) with defined structures that can specifically bind to a molecular target via three-dimensional structures.
-
Similarly to the way antibodies bind to antigens, aptamers specifically recognize and bind to their cognate targets through unique three-dimensional structures.
-
SELEX (systematic evolution of ligands by exponential enrichment) is a gold-standard methodology for generating aptamers, in which an iterative selection procedure — including binding, partitioning, recovery and re-amplification steps — is conducted. Specific sequences (that is, aptamers) can be enriched and dominate the population of library species.
-
Aptamer-based therapeutics typically exploit one of three strategies: an aptamer can serve as an antagonist for blocking the interaction of disease-associated targets (for example, receptor–ligand interactions); an aptamer can serve as an agonist for activating the function of target receptors; or a cell-type-specific aptamer can serve as a carrier for delivering other therapeutic agents to the target cells or tissue.
-
There are three aptamers designated for use in ophthalmology, including one drug approved by the US Food and Drug Administration (FDA) (pegaptanib (Macugen)), and two in late-stage development (ACR-1905 and E-10030).
-
Six RNA and four DNA aptamers have undergone clinical trials for the treatment of various conditions, including macular degeneration, coagulation, oncology and inflammation. All aptamers that have entered clinical trials so far act as antagonists.
Abstract
Nucleic acid aptamers, often termed 'chemical antibodies', are functionally comparable to traditional antibodies, but offer several advantages, including their relatively small physical size, flexible structure, quick chemical production, versatile chemical modification, high stability and lack of immunogenicity. In addition, many aptamers are internalized upon binding to cellular receptors, making them useful targeted delivery agents for small interfering RNAs (siRNAs), microRNAs and conventional drugs. However, several crucial factors have delayed the clinical translation of therapeutic aptamers, such as their inherent physicochemical characteristics and lack of safety data. This Review discusses these challenges, highlighting recent clinical developments and technological advances that have revived the impetus for this promising class of therapeutics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
28 April 2017
Base Pair Biotechnologies and Apterna were omitted from Table 3 in this article. These have now been included.
References
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990). One of the first three publications of the SELEX technology. A related smaller randomized library theoretically containing 48 individual sequences was used for the selection of RNA ligands for T4 DNA polymerase. This procedure was thus named 'systematic evolution of ligands by exponential enrichment' (SELEX).
Robertson, D. L. & Joyce, G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344, 467–468 (1990). One of the first three publications of the SELEX technology. The first RNA enzyme that could cleave ssDNA specifically was selected via in vitro selection.
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990). One of the first three publications of the SELEX technology. The first example of an RNA aptamer specific to small organic dyes.
Mayer, G. The chemical biology of aptamers. Angew. Chem. Int. Ed. 48, 2672–2689 (2009).
Gelinas, A. D., Davies, D. R. & Janjic, N. Embracing proteins: structural themes in aptamer–protein complexes. Curr. Opin. Struct. Biol. 36, 122–132 (2016).
Gold, L. Oligonucleotides as research, diagnostic, and therapeutic agents. J. Biol. Chem. 270, 13581–13584 (1995).
Geiger, A., Burgstaller, P., von der Eltz, H., Roeder, A. & Famulok, M. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 24, 1029–1036 (1996).
Jenison, R. D., Gill, S. C., Pardi, A. & Polisky, B. High-resolution molecular discrimination by RNA. Science 263, 1425–1429 (1994).
Sassanfar, M. & Szostak, J. W. An RNA motif that binds ATP. Nature 364, 550–553 (1993).
Chen, L. et al. The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation. Proc. Natl Acad. Sci. USA 112, 10002–10007 (2015). The first published RNA aptamer specific to a single-amino-acid mutation.
Keefe, A. D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 9, 537–550 (2010).
Sundaram, P., Kurniawan, H., Byrne, M. E. & Wower, J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 48, 259–271 (2013).
Ecker, D. M., Jones, S. D. & Levine, H. L. The therapeutic monoclonal antibody market. MAbs 7, 9–14 (2015).
Ng, E. W. et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 5, 123–132 (2006).
Siddiqui, M. A. & Keating, G. M. Pegaptanib: in exudative age-related macular degeneration. Drugs 65, 1571–1577; discussion 1578–1579 (2005).
Mousa, S. A. & Mousa, S. S. Current status of vascular endothelial growth factor inhibition in age-related macular degeneration. BioDrugs 24, 183–194 (2010).
Ferrara, N. & Adamis, A. P. Ten years of anti-vascular endothelial growth factor therapy. Nat. Rev. Drug Discov. 15 385–403 (2016).
Lincoff, A. M. et al. Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): a randomised clinical trial. Lancet 387, 349–356 (2016). A recent update of the REG1 anticoagulation system in a randomized clinical trial. The severe allergic reactions observed in some patients have been linked to pre-existing antibodies against PEG.
Verheugt, F. W. An anticoagulant too good to be true for revascularisation. Lancet 387, 314–315 (2016).
Bock, L. C., Griffin, L. C., Latham, J. A., Vermaas, E. H. & Toole, J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564–566 (1992). The first DNA aptamers targeting the protease thrombin of the blood coagulation cascade, identified through in vitro SELEX.
Shu, Y. et al. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv. Drug Deliv. Rev. 66, 74–89 (2014).
Kulbachinskiy, A. V. Methods for selection of aptamers to protein targets. Biochemistry (Mosc.) 72, 1505–1518 (2007).
Ozer, A., Pagano, J. M. & Lis, J. T. New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol. Ther. Nucleic Acids 3, e183 (2014).
Keefe, A. D. & Cload, S. T. SELEX with modified nucleotides. Curr. Opin. Chem. Biol. 12, 448–456 (2008).
Pieken, W. A., Olsen, D. B., Benseler, F., Aurup, H. & Eckstein, F. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science 253, 314–317 (1991).
Nimjee, S. M., Rusconi, C. P. & Sullenger, B. A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 56, 555–583 (2005).
Pestourie, C. et al. Comparison of different strategies to select aptamers against a transmembrane protein target. Oligonucleotides 16, 323–335 (2006).
Cerchia, L. & de Franciscis, V. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 28, 517–525 (2010).
Cerchia, L. et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol. 3, e123 (2005).
Liu, Y. et al. Aptamers selected against the unglycosylated EGFRvIII ectodomain and delivered intracellularly reduce membrane-bound EGFRvIII and induce apoptosis. Biol. Chem. 390, 137–144 (2009).
Guo, K. T., Paul, A., Schichor, C., Ziemer, G. & Wendel, H. P. Cell-SELEX: novel perspectives of aptamer-based therapeutics. Int. J. Mol. Sci. 9, 668–678 (2008).
Ohuchi, S. Cell-SELEX technology. Biores Open Access 1, 265–272 (2012).
Cerchia, L., Giangrande, P. H., McNamara, J. O. & de Franciscis, V. Cell-specific aptamers for targeted therapies. Methods Mol. Biol. 535, 59–78 (2009).
Tang, Z. et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 79, 4900–4907 (2007).
Xiao, Z., Shangguan, D., Cao, Z., Fang, X. & Tan, W. Cell-specific internalization study of an aptamer from whole cell selection. Chemistry 14, 1769–1775 (2008).
Phillips, J. A., Lopez-Colon, D., Zhu, Z., Xu, Y. & Tan, W. Applications of aptamers in cancer cell biology. Anal. Chim. Acta 621, 101–108 (2008).
Avci-Adali, M., Metzger, M., Perle, N., Ziemer, G. & Wendel, H. P. Pitfalls of cell-systematic evolution of ligands by exponential enrichment (SELEX): existing dead cells during in vitro selection anticipate the enrichment of specific aptamers. Oligonucleotides 20, 317–323 (2010).
Raddatz, M. S. et al. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew. Chem. Int. Ed. 47, 5190–5193 (2008).
Healy, J. M. et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 21, 2234–2246 (2004).
Mi, J. et al. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 6, 22–24 (2010). The first published RNA aptamers in vivo isolated from mice bearing liver tumours. The target protein of the selected aptamer is p68, an RNA helicase.
Cheng, C., Chen, Y. H., Lennox, K. A., Behlke, M. A. & Davidson, B. L. In vivo SELEX for identification of brain-penetrating aptamers. Mol. Ther. Nucleic Acids 2, e67 (2013). The first brain-penetrating RNA aptamer to be identified via in vivo SELEX.
Darmostuk, M., Rimpelova, S., Gbelcova, H. & Ruml, T. Current approaches in SELEX: an update to aptamer selection technology. Biotechnol. Adv. 33, 1141–1161 (2015).
Ellington, A. D. & Szostak, J. W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355, 850–852 (1992).
Drabovich, A. P., Berezovski, M., Okhonin, V. & Krylov, S. N. Selection of smart aptamers by methods of kinetic capillary electrophoresis. Anal. Chem. 78, 3171–3178 (2006).
Yufa, R. et al. Emulsion PCR significantly improves nonequilibrium capillary electrophoresis of equilibrium mixtures-based aptamer selection: allowing for efficient and rapid selection of aptamer to unmodified ABH2 protein. Anal. Chem. 87, 1411–1419 (2015).
Mosing, R. K., Mendonsa, S. D. & Bowser, M. T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 77, 6107–6112 (2005).
Miyachi, Y., Shimizu, N., Ogino, C. & Kondo, A. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 38, e21 (2010).
Peng, L., Stephens, B. J., Bonin, K., Cubicciotti, R. & Guthold, M. A combined atomic force/fluorescence microscopy technique to select aptamers in a single cycle from a small pool of random oligonucleotides. Microsc. Res. Tech. 70, 372–381 (2007).
Mayer, G. et al. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 5, 1993–2004 (2010).
Huang, C. J., Lin, H. I., Shiesh, S. C. & Lee, G. B. Integrated microfluidic system for rapid screening of CRP aptamers utilizing systematic evolution of ligands by exponential enrichment (SELEX). Biosens. Bioelectron. 25, 1761–1766 (2010).
Cho, M. et al. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl Acad. Sci. USA 107, 15373–15378 (2010).
Sayer, N., Ibrahim, J., Turner, K., Tahiri-Alaoui, A. & James, W. Structural characterization of a 2'F-RNA aptamer that binds a HIV-1 SU glycoprotein, gp120. Biochem. Biophys. Res. Commun. 293, 924–931 (2002).
Kanagawa, T. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96, 317–323 (2003).
Musheev, M. U. & Krylov, S. N. Selection of aptamers by systematic evolution of ligands by exponential enrichment: addressing the polymerase chain reaction issue. Anal. Chim. Acta 564, 91–96 (2006).
Schutze, T. et al. A streamlined protocol for emulsion polymerase chain reaction and subsequent purification. Anal. Biochem. 410, 155–157 (2011).
Shao, K. et al. Emulsion PCR: a high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS ONE 6, e24910 (2011).
Nakano, M. et al. Single-molecule PCR using water-in-oil emulsion. J. Biotechnol. 102, 117–124 (2003).
Ouellet, E., Foley, J. H., Conway, E. M. & Haynes, C. Hi-fi SELEX: a high-fidelity digital-PCR based therapeutic aptamer discovery platform. Biotechnol. Bioeng. 112, 1506–1522 (2015).
Levay, A. et al. Identifying high-affinity aptamer ligands with defined cross-reactivity using high-throughput guided systematic evolution of ligands by exponential enrichment. Nucleic Acids Res. 43, e82 (2015).
Hoon, S., Zhou, B., Janda, K. D., Brenner, S. & Scolnick, J. Aptamer selection by high-throughput sequencing and informatic analysis. BioTechniques 51, 413–416 (2011).
Hoinka, J. et al. Large scale analysis of the mutational landscape in HT-SELEX improves aptamer discovery. Nucleic Acids Res. 43, 5699–5707 (2015).
Thiel, W. H. et al. Nucleotide bias observed with a short SELEX RNA aptamer library. Nucleic Acid. Ther. 21, 253–263 (2011).
Thiel, W. H. et al. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS ONE 7, e43836 (2012).
Kuwahara, M. & Sugimoto, N. Molecular evolution of functional nucleic acids with chemical modifications. Molecules 15, 5423–5444 (2010).
Jager, S. et al. A versatile toolbox for variable DNA functionalization at high density. J. Am. Chem. Soc. 127, 15071–15082 (2005).
Hirao, I. et al. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods 3, 729–735 (2006).
Lin, Y., Qiu, Q., Gill, S. C. & Jayasena, S. D. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res. 22, 5229–5234 (1994).
Ruckman, J. et al. 2′-fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 273, 20556–20567 (1998).
Burmeister, P. E. et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem. Biol. 12, 25–33 (2005).
Kuwahara, M. & Obika, S. In vitro selection of BNA (LNA) aptamers. Artif. DNA PNA XNA 4, 39–48 (2013).
Veedu, R. N. & Wengel, J. Locked nucleic acid nucleoside triphosphates and polymerases: on the way towards evolution of LNA aptamers. Mol. Biosyst. 5, 787–792 (2009).
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 5, e15004 (2010).
Vaught, J. D. et al. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc. 132, 4141–4151 (2010).
Maasch, C., Buchner, K., Eulberg, D., Vonhoff, S. & Klussmann, S. Physicochemical stability of NOX-E36, a 40mer l-RNA (Spiegelmer) for therapeutic applications. Nucleic Acids Symp. Ser. (Oxf.) 52, 61–62 (2008).
Vater, A. & Klussmann, S. Toward third-generation aptamers: Spiegelmers and their therapeutic prospects. Curr. Opin. Drug Discov. Devel. 6, 253–261 (2003).
Lee, Y., Urban, J. H., Xu, L., Sullenger, B. A. & Lee, J. 2′fluoro modification differentially modulates the ability of RNAs to activate pattern recognition receptors. Nucleic Acid Ther. 26, 173–182 (2016). RNAs containing 2′-fluoropyrimidines differentially controlled the activation of pattern recognition receptors. The results demonstrate that RNAs containing 2′-fluoropyrimidine and 5′-triphosphate increased cell death and IFNβ expression in human cancer cells.
Aaldering, L. J. et al. Smart functional nucleic acid chimeras: enabling tissue specific RNA targeting therapy. RNA Biol. 12, 412–425 (2015).
Guo, P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 5, 833–842 (2010).
Rusconi, C. P. et al. Antidote-mediated control of an anticoagulant aptamer in vivo. Nat. Biotechnol. 22, 1423–1428 (2004).
Lee, C. H. et al. Pharmacokinetics of a cholesterol-conjugated aptamer against the hepatitis C virus (HCV) NS5B protein. Mol. Ther. Nucleic Acids 4, e254 (2015).
Dougan, H. et al. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl. Med. Biol. 27, 289–297 (2000).
Heo, K. et al. An aptamer-antibody complex (oligobody) as a novel delivery platform for targeted cancer therapies. J. Control. Release 229, 1–9 (2016).
Willis, M. C. et al. Liposome-anchored vascular endothelial growth factor aptamers. Bioconjug. Chem. 9, 573–582 (1998).
Zhou, J. et al. A hybrid DNA aptamer-dendrimer nanomaterial for targeted cell labeling. Macromol. Biosci. 9, 831–835 (2009).
Musumeci, D. & Montesarchio, D. Polyvalent nucleic acid aptamers and modulation of their activity: a focus on the thrombin binding aptamer. Pharmacol. Ther. 136, 202–215 (2012).
Soule, E. E., Bompiani, K. M., Woodruff, R. S. & Sullenger, B. A. Targeting two coagulation cascade proteases with a bivalent aptamer yields a potent and antidote-controllable anticoagulant. Nucleic Acid Ther. 26, 1–9 (2016).
Kim, Y., Dennis, D. M., Morey, T., Yang, L. & Tan, W. Engineering dendritic aptamer assemblies as superior inhibitors of protein function. Chem. Asian J. 5, 56–59 (2010).
Drolet, D. W. et al. Pharmacokinetics and safety of an anti-vascular endothelial growth factor aptamer (NX1838) following injection into the vitreous humor of rhesus monkeys. Pharm. Res. 17, 1503–1510 (2000).
Tucker, C. E. et al. Detection and plasma pharmacokinetics of an anti-vascular endothelial growth factor oligonucleotide-aptamer (NX1838) in rhesus monkeys. J. Chromatogr. B Biomed. Sci. Appl. 732, 203–212 (1999).
Borbas, K. E., Ferreira, C. S., Perkins, A., Bruce, J. I. & Missailidis, S. Design and synthesis of mono- and multimeric targeted radiopharmaceuticals based on novel cyclen ligands coupled to anti-MUC1 aptamers for the diagnostic imaging and targeted radiotherapy of cancer. Bioconjug. Chem. 18, 1205–1212 (2007).
Choi, D. Y. et al. Sustained elevated intraocular pressures after intravitreal injection of bevacizumab, ranibizumab, and pegaptanib. Retina 31, 1028–1035 (2011).
Steffensmeier, A. C., Azar, A. E., Fuller, J. J., Muller, B. A. & Russell, S. R. Vitreous injections of pegaptanib sodium triggering allergic reactions. Am. J. Ophthalmol. 143, 512–513 (2007).
Agrawal, S., Joshi, M. & Christoforidis, J. B. Vitreous inflammation associated with intravitreal anti-VEGF pharmacotherapy. Mediators Inflamm. 2013, 943409 (2013).
Falavarjani, K. G. & Nguyen, Q. D. Adverse events and complications associated with intravitreal injection of anti-VEGF agents: a review of literature. Eye (Lond.) 27, 787–794 (2013).
Boyer, D. S., Goldbaum, M., Leys, A. M., Starita, C. & V.I.S.I.O.N. Study Group. Effect of pegaptanib sodium 0.3 mg intravitreal injections (Macugen) in intraocular pressure: posthoc analysis from V.I.S.I.O.N. study. Br. J. Ophthalmol. 98, 1543–1546 (2014).
Henry, S. P. et al. Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: potential mechanism of action. J. Pharmacol. Exp. Ther. 281, 810–816 (1997).
Farman, C. A. & Kornbrust, D. J. Oligodeoxynucleotide studies in primates: antisense and immune stimulatory indications. Toxicol. Pathol. 31, S119–S122 (2003).
Swayze, E. E. et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35, 687–700 (2007).
Ganson, N. J. et al. Pre-existing anti-polyethylene glycol antibody linked to first-exposure allergic reactions to pegnivacogin, a PEGylated RNA aptamer. J. Allergy Clin. Immunol. 137, 1610–1613.e7 (2016).
Waring, M. J. Lipophilicity in drug discovery. Expert Opin. Drug Discov. 5, 235–248 (2010).
Zhou, J. & Rossi, J. J. Cell-type-specific, aptamer-functionalized agents for targeted disease therapy. Mol. Ther. Nucleic Acids 3, e169 (2014).
Shum, K. T., Zhou, J. & Rossi, J. J. Aptamer-based therapeutics: new approaches to combat human viral diseases. Pharmaceuticals (Basel) 6, 1507–1542 (2013).
Kohn, D. B. et al. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34+ cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood 94, 368–371 (1999).
DiGiusto, D. L. et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34+ cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl Med. 2, 36ra43 (2010).
Michienzi, A., Li, S., Zaia, J. A. & Rossi, J. J. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc. Natl Acad. Sci. USA 99, 14047–14052 (2002).
Unwalla, H. J. & Rossi, J. J. A dual function TAR decoy serves as an anti-HIV siRNA delivery vehicle. Virol. J. 7, 33 (2010).
Drolet, D. W., Green, L. S., Gold, L. & Janjic, N. Fit for the eye: aptamers in ocular disorders. Nucleic Acid Ther. 26, 127–146 (2016). A comprehensive review about the development and current progress of three aptamers in ophthalmology.
Eyetech Study Group. Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina 22, 143–152 (2002).
Bullock, A. N. & Fersht, A. R. Rescuing the function of mutant p53. Nat. Rev. Cancer 1, 68–76 (2001).
Axel, D. I. et al. Paclitaxel inhibits arterial smooth muscle cell proliferation and migration in vitro and in vivo using local drug delivery. Circulation 96, 636–645 (1997).
Thiel, W. H. et al. Smooth muscle cell-targeted RNA aptamer inhibits neointimal formation. Mol. Ther. 24, 779–787 (2016).
Nimjee, S. M., Rusconi, C. P., Harrington, R. A. & Sullenger, B. A. The potential of aptamers as anticoagulants. Trends Cardiovasc. Med. 15, 41–45 (2005).
Bompiani, K. M. et al. Probing the coagulation pathway with aptamers identifies combinations that synergistically inhibit blood clot formation. Chem. Biol. 21, 935–944 (2014).
Nimjee, S. M. et al. Synergistic effect of aptamers that inhibit exosites 1 and 2 on thrombin. RNA 15, 2105–2111 (2009).
Dyke, C. K. et al. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation 114, 2490–2497 (2006).
Chen, C. H., Chernis, G. A., Hoang, V. Q. & Landgraf, R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl Acad. Sci. USA 100, 9226–9231 (2003).
Dollins, C. M. et al. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 15, 675–682 (2008). One of the first two studies using aptamers as agonists. A bivalent aptamer was created to achieve an agonistic effect.
Pratico, E. D., Sullenger, B. A. & Nair, S. K. Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic Acid Ther. 23, 35–43 (2013).
McNamara, J. O. et al. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Invest. 118, 376–386 (2008). One of the first two studies using aptamers as agonists. A bivalent aptamer was created to achieve an agonistic effect.
Soldevilla, M. M. et al. 2-Fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia. Biomaterials 67, 274–285 (2015).
Pastor, F. et al. CD28 aptamers as powerful immune response modulators. Mol. Ther. Nucleic Acids 2, e98 (2013).
Ramaswamy, V. et al. DNA aptamer assembly as a vascular endothelial growth factor receptor agonist. Nucleic Acid. Ther. 25, 227–234 (2015).
Yunn, N. O. et al. Agonistic aptamer to the insulin receptor leads to biased signaling and functional selectivity through allosteric modulation. Nucleic Acids Res. 43, 7688–7701 (2015).
Gilboa, E., McNamara, J. & Pastor, F. Use of oligonucleotide aptamer ligands to modulate the function of immune receptors. Clin. Cancer Res. 19, 1054–1062 (2013).
Khedri, M., Rafatpanah, H., Abnous, K., Ramezani, P. & Ramezani, M. Cancer immunotherapy via nucleic acid aptamers. Int. Immunopharmacol. 29, 926–936 (2015).
Compaan, D. M. & Hymowitz, S. G. The crystal structure of the costimulatory OX40-OX40L complex. Structure 14, 1321–1330 (2006).
Tasch, J., Gong, M., Sadelain, M. & Heston, W. D. A unique folate hydrolase, prostate-specific membrane antigen (PSMA): a target for immunotherapy? Crit. Rev. Immunol. 21, 249–261 (2001).
McNamara, J. O. et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 24, 1005–1015 (2006). One of the first two studies using PSMA aptamers for siRNA delivery.
Dassie, J. P. et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 27, 839–849 (2009).
Pastor, F., Kolonias, D., Giangrande, P. H. & Gilboa, E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 465, 227–230 (2010).
Ni, X. et al. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J. Clin. Invest. 121, 2383–2390 (2011).
Wullner, U. et al. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr. Cancer Drug Targets 8, 554–565 (2008).
Chu, T. C., Twu, K. Y., Ellington, A. D. & Levy, M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 34, e73 (2006).
Pastor, F., Kolonias, D., McNamara, J. O. II & Gilboa, E. Targeting 4-1BB costimulation to disseminated tumor lesions with bi-specific oligonucleotide aptamers. Mol. Ther. 19, 1878–1886 (2011).
Zhou, J. et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 37, 3094–3109 (2009).
Zhou, J., Li, H., Li, S., Zaia, J. & Rossi, J. J. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 16, 1481–1489 (2008).
Zhou, J. et al. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther. 21, 192–200 (2013).
Neff, C. P. et al. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4+ T cell decline in humanized mice. Sci. Transl Med. 3, 66ra6 (2011).
Catuogno, S., Rienzo, A., Di Vito, A., Esposito, C. L. & de Franciscis, V. Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J. Control. Release 210, 147–159 (2015).
Shu, D. et al. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano 9, 9731–9740 (2015).
Chu, T. C. et al. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 66, 5989–5992 (2006). The first study using PSMA aptamers for toxin delivery.
Hernandez, L. I. et al. Methods for evaluating cell-specific, cell-internalizing RNA aptamers. Pharmaceuticals (Basel) 6, 295–319 (2013).
Boyacioglu, O., Stuart, C. H., Kulik, G. & Gmeiner, W. H. Dimeric DNA aptamer complexes for high-capacity-targeted drug delivery using pH-sensitive covalent linkages. Mol. Ther. Nucleic Acids 2, e107 (2013).
Bagalkot, V., Farokhzad, O. C., Langer, R. & Jon, S. An aptamer–doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. 45, 8149–8152 (2006).
Gijs, M., Aerts, A., Impens, N., Baatout, S. & Luxen, A. Aptamers as radiopharmaceuticals for nuclear imaging and therapy. Nucl. Med. Biol. 43, 253–271 (2016).
Younes, C. K., Boisgard, R. & Tavitian, B. Labelled oligonucleotides as radiopharmaceuticals: pitfalls, problems and perspectives. Curr. Pharm. Des. 8, 1451–1466 (2002).
Sugiura, G., Kuhn, H., Sauter, M., Haberkorn, U. & Mier, W. Radiolabeling strategies for tumor-targeting proteinaceous drugs. Molecules 19, 2135–2165 (2014).
Hicke, B. J. et al. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 276, 48644–48654 (2001).
Hicke, B. J. et al. Tumor targeting by an aptamer. J. Nucl. Med. 47, 668–678 (2006).
Lao, Y. H., Phua, K. K. & Leong, K. W. Aptamer nanomedicine for cancer therapeutics: barriers and potential for translation. ACS Nano 9, 2235–2254 (2015).
Farokhzad, O. C. et al. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 64, 7668–7672 (2004).
Liang, C. et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat. Med. 21, 288–294 (2015).
Barakat, M. R. & Kaiser, P. K. VEGF inhibitors for the treatment of neovascular age-related macular degeneration. Expert Opin. Investig. Drugs 18, 637–646 (2009).
VEGF Inhibition Study in Oracular Neovascularization (V. I. S. I. O. N.) Clinical Trial Group et al. Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology 113, 1508.e1–1508.e25 (2006).
Jellinek, D., Lynott, C. K., Rifkin, D. B. & Janjic, N. High-affinity RNA ligands to basic fibroblast growth factor inhibit receptor binding. Proc. Natl Acad. Sci. USA 90, 11227–11231 (1993).
Shaw, J. P., Kent, K., Bird, J., Fishback, J. & Froehler, B. Modified deoxyoligonucleotides stable to exonuclease degradation in serum. Nucleic Acids Res. 19, 747–750 (1991).
Jellinek, D. et al. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 34, 11363–11372 (1995).
Jaschke, A. et al. Synthesis and properties of oligodeoxyribonucleotide-polyethylene glycol conjugates. Nucleic Acids Res. 22, 4810–4817 (1994).
Kawaguchi, T., Asakawa, H., Tashiro, Y., Juni, K. & Sueishi, T. Stability, specific binding activity, and plasma concentration in mice of an oligodeoxynucleotide modified at 5′-terminal with poly(ethylene glycol). Biol. Pharm. Bull. 18, 474–476 (1995).
CATT Research Group et al. Ranibizumab and bevacizumab for neovascular age-related macular degeneration. N. Engl. J. Med. 364, 1897–1908 (2011).
Brown, D. M. et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N. Engl. J. Med. 355, 1432–1444 (2006).
Rosenfeld, P. J. et al. Ranibizumab for neovascular age-related macular degeneration. N. Engl. J. Med. 355, 1419–1431 (2006).
Heier, J. S. et al. Intravitreal aflibercept (VEGF trap-eye) in wet age-related macular degeneration. Ophthalmology 119, 2537–2548 (2012).
D'Amore, P. A. Vascular endothelial cell growth factor-a: not just for endothelial cells anymore. Am. J. Pathol. 171, 14–18 (2007).
Ratanji, K. D., Derrick, J. P., Dearman, R. J. & Kimber, I. Immunogenicity of therapeutic proteins: influence of aggregation. J. Immunotoxicol. 11, 99–109 (2014).
Kawa, M. P., Machalinska, A., Roginska, D. & Machalinski, B. Complement system in pathogenesis of AMD: dual player in degeneration and protection of retinal tissue. J. Immunol. Res. 2014, 483960 (2014).
Baas, D. C. et al. The complement component 5 gene and age-related macular degeneration. Ophthalmology 117, 500–511 (2010).
Biesecker, G., Dihel, L., Enney, K. & Bendele, R. A. Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology 42, 219–230 (1999).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT02397954?term=NCT02397954&rank=1 (2016).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT02686658?term=NCT02686658&rank=1 (2016).
Sadiq, M. A. et al. Platelet derived growth factor inhibitors: a potential therapeutic approach for ocular neovascularization. Saudi J. Ophthalmol. 29, 287–291 (2015).
Mabry, R. et al. A dual-targeting PDGFRβ/VEGF-A molecule assembled from stable antibody fragments demonstrates anti-angiogenic activity in vitro and in vivo. MAbs 2, 20–34 (2010).
Jo, N. et al. Inhibition of platelet-derived growth factor B signaling enhances the efficacy of anti-vascular endothelial growth factor therapy in multiple models of ocular neovascularization. Am. J. Pathol. 168, 2036–2053 (2006).
Boyer, D. S. Phase 2b study of Fovista™, a platelet derived growth factor (PDGF) inhibitor in combination with a vascular endothelial growth factor (VEGF) inhibitor for neovascular age-related macular degeneration (AMD). Invest. Ophthalmol. Vis. Sci. 54, abstr. 2175 (2013).
Boyer, D. S. Combined inhibition of platelet derived (PDGF) and vascular endothelial (VEGF) growth factors for the treatment of neovascular age-related macular degeration (NV-AMD) - results of a Phase I study. Invest. Ophthalmol. Vis. Sci. 50, abstr. 1260 (2009).
Green, L. S. et al. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry 35, 14413–14424 (1996).
Floege, J. et al. Novel approach to specific growth factor inhibition in vivo: antagonism of platelet-derived growth factor in glomerulonephritis by aptamers. Am. J. Pathol. 154, 169–179 (1999).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01089517?term=NCT01089517&rank=1 (2013).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01944839?term=NCT01944839&rank=1 (2005).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01940900?term=NCT01940900&rank=1 (2015).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01940887?term=NCT01940887&rank=1 (2016).
Smiley, D. A. & Becker, R. C. Factor IXa as a target for anticoagulation in thrombotic disorders and conditions. Drug Discov. Today 19, 1445–1453 (2014).
Rusconi, C. P. et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419, 90–94 (2002).
Povsic, T. J. et al. Use of the REG1 anticoagulation system in patients with acute coronary syndromes undergoing percutaneous coronary intervention: results from the phase II RADAR-PCI study. EuroIntervention 10, 431–438 (2014).
Povsic, T. J. et al. A Phase 2, randomized, partially blinded, active-controlled study assessing the efficacy and safety of variable anticoagulation reversal using the REG1 system in patients with acute coronary syndromes: results of the RADAR trial. Eur. Heart J. 34, 2481–2489 (2013).
Povsic, T. J. et al. Pegnivacogin results in near complete FIX inhibition in acute coronary syndrome patients: RADAR pharmacokinetic and pharmacodynamic substudy. Eur. Heart J. 32, 2412–2419 (2011).
Cohen, M. G. et al. First clinical application of an actively reversible direct factor IXa inhibitor as an anticoagulation strategy in patients undergoing percutaneous coronary intervention. Circulation 122, 614–622 (2010).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00932100?term=NCT00932100&rank=1 (2012).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00715455?term=NCT00715455&rank=1 (2013).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01872572?term=NCT01872572&rank=1 (2013).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01848106?term=NCT01848106&rank=1 (2014).
Diener, J. L. et al. Inhibition of von Willebrand factor-mediated platelet activation and thrombosis by the anti-von Willebrand factor A1-domain aptamer ARC1779. J. Thromb. Haemost. 7, 1155–1162 (2009).
Jilma, B. et al. A randomised pilot trial of the anti-von Willebrand factor aptamer ARC1779 in patients with type 2b von Willebrand disease. Thromb. Haemost. 104, 563–570 (2010).
Gilbert, J. C. et al. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation 116, 2678–2686 (2007).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00432770?term=ARC1779&rank=6 (2007).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00632242?term=ARC1779&rank=1 (2009).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00742612?term=ARC1779&rank=2 (2010).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00507338?term=ARC1779&rank=5 (2009).
Waters, E. K., Richardson, J., Schaub, R. G. & Kurz, J. C. Effect of NU172 and bivalirudin on ecarin clotting time in human plasma and whole blood. J. Thromb. Haemost. 7, 683 (2009).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00808964?term=00808964&rank=1 (2011).
Waters, E. K. et al. Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood 117, 5514–5522 (2011).
Gorczyca, M. E. et al. Inhibition of tissue factor pathway inhibitor (TFPI) by ARC19499 improves clotting of hemophiliac blood. BMC Pharmacol. 10, abstr. A44 (2010).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01191372?term=NCT01191372&rank=1 (2015).
Soundararajan, S. et al. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol. Pharmacol. 76, 984–991 (2009).
Bates, P. J., Kahlon, J. B., Thomas, S. D., Trent, J. O. & Miller, D. M. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 274, 26369–26377 (1999).
Soundararajan, S., Chen, W., Spicer, E. K., Courtenay-Luck, N. & Fernandes, D. J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 68, 2358–2365 (2008).
Berger, C. M., Gaume, X. & Bouvet, P. The roles of nucleolin subcellular localization in cancer. Biochimie 113, 78–85 (2015).
Mongelard, F. & Bouvet, P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr. Opin. Mol. Ther. 12, 107–114 (2010).
Reyes-Reyes, E. M., Teng, Y. & Bates, P. J. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 70, 8617–8629 (2010).
Bates, P. J., Laber, D. A., Miller, D. M., Thomas, S. D. & Trent, J. O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 86, 151–164 (2009).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00881244?term=NCT00881244&rank=1 (2009).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00512083?term=NCT00512083&rank=1 (2009).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00740441?term=NCT00740441&rank=1 (2009).
Hoellenriegel, J. et al. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood 123, 1032–1039 (2014).
Marasca, R. & Maffei, R. NOX-A12: mobilizing CLL away from home. Blood 123, 952–953 (2014).
Liu, S. C. et al. Blockade of SDF-1 after irradiation inhibits tumor recurrences of autochthonous brain tumors in rats. Neuro Oncol. 16, 21–28 (2014).
Sayyed, S. G. et al. Podocytes produce homeostatic chemokine stromal cell-derived factor-1/CXCL12, which contributes to glomerulosclerosis, podocyte loss and albuminuria in a mouse model of type 2 diabetes. Diabetologia 52, 2445–2454 (2009).
Burns, J. M. et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J. Exp. Med. 203, 2201–2213 (2006).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00976378?term=NCT00976378&rank=1 (2014).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01194934?term=NCT01194934&rank=1 (2014).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01486797?term=NCT01486797&rank=1 (2016).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01521533?term=NCT01521533&rank=1 (2015).
Oberthur, D. et al. Crystal structure of a mirror-image l-RNA aptamer (Spiegelmer) in complex with the natural l-protein target CCL2. Nat. Commun. 6, 6923 (2015).
Ninichuk, V. et al. Late onset of Ccl2 blockade with the Spiegelmer mNOX-E36-3'PEG prevents glomerulosclerosis and improves glomerular filtration rate in db/db mice. Am. J. Pathol. 172, 628–637 (2008).
Kulkarni, O. et al. Spiegelmer inhibition of CCL2/MCP-1 ameliorates lupus nephritis in MRL-(Fas)lpr mice. J. Am. Soc. Nephrol. 18, 2350–2358 (2007).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01085292?term=NCT01085292&rank=1 (2013).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01547897?term=NCT01547897&rank=1 (2014).
Schwoebel, F. et al. The effects of the anti-hepcidin Spiegelmer NOX-H94 on inflammation-induced anemia in cynomolgus monkeys. Blood 121, 2311–2315 (2013).
Ganz, T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood 102, 783–788 (2003).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01372137?term=NCT01372137&rank=1 (2016).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01691040?term=NCT01691040&rank=1 (2014).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT02079896?term=NCT02079896&rank=1 (2015).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00021736?term=NCT00021736&rank=1 (2005).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00040313?term=00040313&rank=1 (2006).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00056199?term=00056199&rank=1 (2008).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00312351?term=00312351&rank=1 (2007).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00321997?term=NCT00321997&rank=1 (2006).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01487070?term=NCT01487070&rank=1 (2011).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00709527?term=NCT00709527&rank=1 (2012).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00950638?term=NCT00950638&rank=1 (2013).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00569140?term=NCT00569140&rank=1 (2010).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT02387957?term=NCT02387957&rank=1 (2016).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT02591914?term=NCT02591914&rank=1 (2005).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00113997?term=NCT00113997&rank=1 (2008).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00694785?term=ARC1779&rank=3 (2009).
Jilma-Stohlawetz, P. et al. Inhibition of von Willebrand factor by ARC1779 in patients with acute thrombotic thrombocytopenic purpura. Thromb. Haemost. 105, 545–552 (2011).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01034410?term=NCT01034410&rank=1 (2011).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT00976729?term=NCT00976729&rank=1 (2013).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01372124?term=NCT01372124&rank=1 (2012).
US National Library of Medicine. Clinicaltrials.govhttps://clinicaltrials.gov/ct2/show/NCT01522794?term=NCT01522794&rank=1 (2014).
Acknowledgements
This work was supported by the US National Institutes of Health (grant numbers R01AI29329, R01AI42552 and R01HL07470 to J.J.R.). Funding for open access charge was provided by the US National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the US National Institutes of Health. The authors thank S. T. Wilkinson (City of Hope) for helpful advice in scientific writing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.J.R. and J.Z. have an issued patent entitled “Cell-type specific aptamer-siRNA delivery system for HIV-1 therapy” [USPTO, No. US 8, 222, 226 B2, July 17, 2012]. J.J.R., J.Z., Marco S. Weinberg and Kevin V. Morris have a patent pending on “Cell-specific internalizing RNA aptamers against human CCR5 and used therefore” [United States Patent, application number: 62/025, 368, filed on July 16, 2014].
Glossary
- Nucleic acid aptamers
-
Short, single-stranded DNA or RNA molecules (20–100 nucleotides in length) with defined structures that can specifically bind to a molecular target via three-dimensional structures.
- Systematic evolution of ligands by exponential enrichment
-
(SELEX). An iterative selection procedure for aptamer generation. Each cycle consists of binding, partitioning, recovery and re-amplification steps. Specific sequences (that is, aptamers) can be enriched and dominate the population of library species.
- G-quadruplex
-
A G-quadruplex structure can be formed in a guanine-rich sequence when four guanine bases are associated through hydrogen bonding.
- Kissing hairpin
-
If two RNA stem-loops have complementary sequences in the loop regions, the two loops will base pair to form a kissing complex.
- Macugen
-
Trade name of pegaptanib, a modified RNA aptamer that targets vascular endothelial growth factor (VEGF), and the first federally approved aptamer drug for the treatment of wet age-related macular degeneration.
- Vascular endothelial growth factor
-
(VEGF). A secreted protein that is capable of inducing angiogenesis and increasing vascular permeability and inflammation.
- Bevacizumab
-
Trade name Avastin; a recombinant humanized monoclonal antibody against vascular endothelial growth factor (VEGF), and an approved antibody drug for the treatment of certain metastatic cancers and eye diseases.
- Ranibizumab
-
Trade name Lucentis; a monoclonal antibody fragment created from bevacizumab, and an approved antibody drug for the treatment of age-related macular degeneration.
- Hairpin
-
A complementary region of nucleic acid that can form Watson–Crick base pairs and generally results in a stem–loop structure that resembles a hairpin.
- Emulsion PCR (ePCR) or droplet digital PCR (ddPCR)
-
Systems that compartmentalize and miniaturize PCR by generating a water-in-oil emulsion containing numerous droplets, which creates a local homogeneous amplification microenvironment.
- High-throughput sequencing
-
(HTS). A next-generation sequencing technology that is capable of parallelizing the sequencing process and producing thousands or millions of sequences at once.
- SOMAmer
-
(Slow off-rate modified aptamer). A chemically modified DNA aptamer that is highly specific for its respective cognate protein target; selected through an in vitro SELEX procedure, in which modified deoxyuracil carrying hydrophobic functional groups is incorporated. More than 1,300 different SOMAmer agents have been developed by SomaLogic.
- Spiegelmers
-
Trade name of the L-ribonucleic acid aptamers developed by NOXXON Pharma; mirror-image RNAs built from natural L-ribose units.
- REG1 system
-
A modified RNA aptamer RB006 (pegnivacogin) and an antidote oligonucleotide RB007 (anivamersen); the first aptamer-based anticoagulation system in clinical trials.
- Co-stimulatory receptors
-
A class of molecules expressed by T lymphocytes that regulate the activation of T cells and the generation of effector T cell responses, including OX40, 4-1BB, CD40, CD28 and programmed cell death protein 1 (PD1). OX40, 4-1BB and CD40 belong to the tumour necrosis factor (TNF) family and are involved in the later phase of T cell activation, whereas CD28 is a member of the larger immunoglobulin superfamily and involved in triggering of the cell-mediated immune response.
- Short hairpin RNA
-
(shRNA). An artificial RNA molecule with a short hairpin turn; like small interfering RNAs (siRNAs), this is another class of an RNA interference (RNAi) trigger.
- Nonsense-mediated mRNA decay
-
(NMD). A translation-coupled mechanism that degrades mRNA containing premature translation-termination codons (PTCs).
- Small interfering RNAs
-
(siRNAs). A class of double-stranded RNA molecules 20–25 base pairs in length that are capable of triggering sequence-specific, post-transcriptional gene silencing.
- MicroRNAs
-
(miRNAs). A class of small non-coding RNA molecules ∼22 nucleotides in length. The mechanism of miRNA-mediated silencing is repression of target mRNA translation accompanied by deadenylation and subsequent degradation of the mRNA targets.
- AntimiRs
-
Synthetic oligonucleotides designed to neutralize microRNA function.
- RNA interference
-
(RNAi). A highly conserved endogenous process for post-transcriptional regulation of gene silencing that is triggered by small regulatory RNAs, including small interfering RNAs.
- gp120
-
An HIV-1 envelope glycoprotein. gp120 is exposed on the surface of virus particles and the plasma membrane of HIV-1-infected cells. The interaction of HIV-1 gp120 with the cellular CD4 receptor is a crucial step in the entry of HIV into T cells.
- Gelonin
-
A small 28 kDa N-glycosidase protein capable of inducing cell death.
- Plekho1
-
(Pleckstrin homology domain-containing family O member 1). Has a role in the regulation of the actin cytoskeleton through its interactions with actin capping protein.
- ARC-1905
-
A modified RNA aptamer targeting complement 5 (C5) protein that has undergone clinical trials for the treatment of dry age-related macular degeneration.
- E-10030
-
A modified DNA aptamer targeting platelet-derived growth factor (PDGF) that has undergone clinical trials for wet age-related macular degeneration therapy.
- Aflibercept
-
Trade name Eylea; a recombinant fusion protein inhibitor of vascular endothelial growth factor (VEGF), and an approved biopharmaceutical drug for the treatment of wet age-related macular degeneration.
- von Willebrand factor
-
(vWF). A key factor in the coagulation cascade associated with platelet recruitment. A deficiency or impairment of vWF causes von Willebrand disease.
- von Willebrand disease
-
A condition that can cause extended or excessive bleeding.
- Stromal cell-derived factor 1
-
(SDF1; also known as CXCL12). A small cytokine protein that has an important role in tumour proliferation, new blood vessel formation and metastasis.
- Hepcidin
-
A small 2.8 kDa peptide thought to be the central mediator of iron homeostasis.
Rights and permissions
About this article
Cite this article
Zhou, J., Rossi, J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov 16, 181–202 (2017). https://doi.org/10.1038/nrd.2016.199
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2016.199
This article is cited by
-
Aptamers as an approach to targeted cancer therapy
Cancer Cell International (2024)
-
Novel nanotherapeutics for cancer immunotherapy by albumin nanoparticles functionalized with PD-1 and PD-L1 aptamers
Cancer Nanotechnology (2024)
-
RNAi-based drug design: considerations and future directions
Nature Reviews Drug Discovery (2024)
-
Non-Faradaic aptasensor based on NH2-GO/PPy for the detection of 17β-estradiol
Journal of the Iranian Chemical Society (2024)
-
A SYBR Green I-based aptasensor for the label-free, fluorometric, and anti-interference detection of MeHg+
Analytical and Bioanalytical Chemistry (2024)