Key Points
-
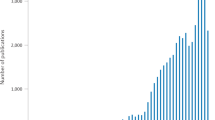
Radiomics is becoming increasingly more important in medical imaging
-
The explosion of medical imaging data creates an environment ideal for machine-learning and data-based science
-
Radiomics-based decision-support systems for precision diagnosis and treatment can be a powerful tool in modern medicine
-
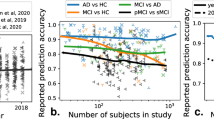
Large-scale data sharing is necessary for the validation and full potential that radiomics represents
-
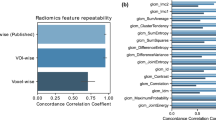
Standardized data collection, evaluation criteria, and reporting guidelines are required for radiomics to mature as a discipline
Abstract
Radiomics, the high-throughput mining of quantitative image features from standard-of-care medical imaging that enables data to be extracted and applied within clinical-decision support systems to improve diagnostic, prognostic, and predictive accuracy, is gaining importance in cancer research. Radiomic analysis exploits sophisticated image analysis tools and the rapid development and validation of medical imaging data that uses image-based signatures for precision diagnosis and treatment, providing a powerful tool in modern medicine. Herein, we describe the process of radiomics, its pitfalls, challenges, opportunities, and its capacity to improve clinical decision making, emphasizing the utility for patients with cancer. Currently, the field of radiomics lacks standardized evaluation of both the scientific integrity and the clinical relevance of the numerous published radiomics investigations resulting from the rapid growth of this area. Rigorous evaluation criteria and reporting guidelines need to be established in order for radiomics to mature as a discipline. Herein, we provide guidance for investigations to meet this urgent need in the field of radiomics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Aerts, H. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Hood, L. & Friend, S. H. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat. Rev. Clin. Oncol. 8, 184–187 (2011).
Lambin, P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 48, 441–446 (2012).
Kumar, V. et al. Radiomics: the process and the challenges. Magn. Reson. Imaging 30, 1234–1248 (2012).
Haase, A. T. et al. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 274, 985–989 (1996).
Lambin, P. et al. Predicting outcomes in radiation oncology — multifactorial decision support systems. Nat. Rev. Clin. Oncol. 10, 27–40 (2013).
[No authors listed] Medicine: Computers by the Bedside. Nature 224, 636–637 (1969).
Schoolman, H. & Bernstein, L. Computer use in diagnosis, prognosis, and therapy. Science 200, 926–931 (1978).
Gillies, R. J., Kinahan, P. E. & Hricak, H. Radiomics: images are more than pictures, they are data. Radiology 278, 563–577 (2016).
Roelofs, E. et al. International data-sharing for radiotherapy research: an open-source based infrastructure for multicentric clinical data mining. Radiother. Oncol. 110, 370–374 (2014).
Roelofs, E. et al. Benefits of a clinical data warehouse with data mining tools to collect data for a radiotherapy trial. Radiother. Oncol. 108, 174–179 (2013).
Miotto, R., Li, L., Kidd, B. A. & Dudley, J. T. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci. Rep. 6, 26094 (2016).
Nead, K. T. et al. Androgen deprivation therapy and future alzheimer's disease risk. J. Clin. Oncol. 34, 566–571 (2016).
Gatenby, R. A., Grove, O. & Gillies, R. J. Quantitative imaging in cancer evolution and ecology. Radiology 269, 8–14 (2013).
Aerts, H. L. The potential of radiomic-based phenotyping in precision medicine: A review. JAMA Oncol. 2, 1636–1642 (2016).
Lambin, P. et al. Decision support systems for personalized and participative radiation oncology. Adv. Drug Delivery Rev. 109, 131–153 (2017).
Vickers, A. Prediction models: revolutionary in principle, but do they do more good than harm? J. Clin. Oncol. 29, 2951–2952 (2011).
Yip, S. S. & Aerts, H. J. Applications and limitations of radiomics. Phys. Med. Biol. 61, R150–166 (2016).
Polan, D. F., Brady, S. L. & Kaufman, R. A. Tissue segmentation of computed tomography images using a Random Forest algorithm: a feasibility study. Phys. Med. Biol. 61, 6553–6569 (2016).
Balagurunathan, Y. et al. Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol. 7, 72–87 (2014).
Grootjans, W. et al. The impact of optimal respiratory gating and image noise on evaluation of intra-tumor heterogeneity in 18F-FDG PET imaging of lung cancer. J. Nucl. Med. 57, 1692–1698 (2016).
Larue, R. T., Defraene, G., de Ruysscher, D., Lambin, P. & van Elmpt, W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br. J. Radiol. 90, 20160665 (2017).
Mackin, D. et al. Measuring computed tomography scanner variability of radiomics features. Invest. Radiol 50, 757–765 (2015).
Balagurunathan, Y. et al. Test–retest reproducibility analysis of lung CT image features. J. Digit. Imaging 27, 805–823 (2014).
Zhao, B. et al. Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non–small cell lung cancer. Radiology 252, 263–272 (2009).
Zhao, B. et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci. Rep. 6, 23428 (2016).
Hatt, M. et al. Characterization of PET/CT images using texture analysis: the past, the present... any future? Eur. J. Nucl. Med. Mol. Imaging 44, 151–165 (2016).
Fang, Y. H. et al. Development and evaluation of an open-source software package “CGITA” for quantifying tumor heterogeneity with molecular images. Biomed. Res. Int. 2014, 248505 (2014).
Zhang, L. et al. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med. Phys. 42, 1341–1353 (2015).
Parmar, C., Grossmann, P., Bussink, J., Lambin, P. & Aerts, H. J. Machine learning methods for quantitative radiomic biomarkers. Sci. Rep. 5, 13087 (2015).
https://github.com/ (2017 May 18 th).
Collins, G., Reitsma, J., Altman, D. & Moons, K. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. Ann. Intern. Med. 162, 55–63 (2015).
Lemeshow, S. & Hosmer, D. W. Jr. A review of goodness of fit statistics for use in the development of logistic regression models. Am. J. Epidemiol. 115, 92–106 (1982).
Debray, T. P. et al. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J. Clin. Epidemiol. 68, 279–289 (2015).
Steyerberg, E. et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 21, 128–138 (2010).
Leek, J. T. & Peng, R. D. Statistics: P values are just the tip of the iceberg. Nature 520, 612 (2015).
Drummond, C. Replicability is not reproducibility: nor is it good science. In Evaluation Methods for Machine Learning (2009).
Peng, R. D. Reproducible research in computational science. Science 334, 1226–1227 (2011).
Peng, R. D., Dominici, F. & Zeger, S. L. Reproducible epidemiologic research. Am. J. Epidemiol. 163, 783–789 (2006).
Lambin, P. Radiomics digital phantom. CancerData.org https://www.cancerdata.org/resource/doi%3A10.17195/candat.2016.08.1 (2017).
http://www.radiomics.world/ (2017 May 18 th).
Altman, D. G., McShane, L. M., Sauerbrei, W. & Taube, S. E. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med. 10, 51 (2012).
Pepe, M. S. & Feng, Z. Improving biomarker identification with better designs and reporting. Clin. Chem. 57, 1093–1095 (2011).
Poste, G. Biospecimens, biomarkers, and burgeoning data: the imperative for more rigorous research standards. Trends Mol. Med. 18, 717–722 (2012).
Rosenstein, B. S. et al. Radiogenomics: radiobiology enters the era of big data and team science. Int. J. Radiat. Oncol. Biol. Phys. 89, 709–713 (2014).
Rutman, A. M. & Kuo, M. D. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur. J. Radiol. 70, 232–241 (2009).
Chang, H. Y. et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl Acad. Sci. USA 102, 3738–3743 (2005).
Chen, X. et al. Gene expression patterns in human liver cancers. Mol. Biol. Cell 13, 1929–1939 (2002).
Chung, C. H., Bernard, P. S. & Perou, C. M. Molecular portraits and the family tree of cancer. Nat. Genet. 32 (Suppl.), 533–540 (2002).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
Paik, S. et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 24, 3726–3734 (2006).
Segal, E., Friedman, N., Kaminski, N., Regev, A. & Koller, D. From signatures to models: understanding cancer using microarrays. Nat. Genet. 37, S38–S45 (2005).
Diehn, M. et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc. Natl Acad. Sci. USA 105, 5213–5218 (2008).
Gevaert, O. et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data—methods and preliminary results. Radiology 264, 387–396 (2012).
Segal, E. et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat. Biotechnol. 25, 675–680 (2007).
Gao, X. et al. The method and efficacy of support vector machine classifiers based on texture features and multi-resolution histogram from 18F-FDG PET-CT images for the evaluation of mediastinal lymph nodes in patients with lung cancer. Eur. J. Radiol. 84, 312–317 (2015).
Harry, V. N., Semple, S. I., Parkin, D. E. & Gilbert, F. J. Use of new imaging techniques to predict tumour response to therapy. Lancet Oncol. 11, 92–102 (2010).
O'Connor, J. P. et al. Quantitative imaging biomarkers in the clinical development of targeted therapeutics: current and future perspectives. Lancet Oncol. 9, 766–776 (2008).
Panth, K. M. et al. Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiother. Oncol. (2015).
Jemal, A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011).
Wang, J. et al. Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: a pilot radiomics study. PLoS ONE 10, e0143308 (2015).
Abernethy, A. et al. Rapid-learning system for cancer care. J. Clin. Oncol. 28, 4268–4274 (2010).
Lambin, P. et al. Modern clinical research: How rapid learning health care and cohort multiple randomised clinical trials complement traditional evidence based medicine. Acta Oncol. 54, 1289–1300 (2015).
Dekker, A. et al. Rapid learning in practice: A lung cancer survival decision support system in routine patient care data. Radiother. Oncol. 113, 47–53 (2014).
Ginsburg, G., Staples, J. & Abernethy, A. Academic medical centers: ripe for rapid-learning personalized health care. Sci. Transl. Med. 3, 101cm27 (2011).
Lambin, P. et al. Rapid learning health care in oncology — An approach towards decision support systems enabling customised radiotherapy. Radiother. Oncol. 109, 159–164 (2013).
Buettner, R., Wolf, J. & Thomas, R. K. Lessons learned from lung cancer genomics: the emerging concept of individualized diagnostics and treatment. J. Clin. Oncol. 31, 1858–1865 (2013).
Colen, R. et al. NCI Workshop Report: clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Transl Oncol. 7, 556–569 (2014).
Rizzo, S. et al. CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer. Eur. Radiol. 26, 32–42 (2015).
Taguchi, F. et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J. Natl Cancer Inst. 99, 838–846 (2007).
Yaromina, A., Krause, M. & Baumann, M. Individualization of cancer treatment from radiotherapy perspective. Mol. Oncol. 6, 211–221 (2012).
Dancey, J. E. et al. Guidelines for the development and incorporation of biomarker studies in early clinical trials of novel agents. Clin. Cancer Res. 16, 1745–1755 (2010).
Krause, M., Yaromina, A., Eicheler, W., Koch, U. & Baumann, M. Cancer stem cells: targets and potential biomarkers for radiotherapy. Clin. Cancer Res. 17, 7224–7229 (2011).
Lindegaard, J. C., Overgaard, J., Bentzen, S. M. & Pedersen, D. Is there a radiobiologic basis for improving the treatment of advanced stage cervical cancer? J. Natl Cancer Inst. Monogr. 21, 105–112 (1996).
Yaromina, A. et al. Pre-treatment number of clonogenic cells and their radiosensitivity are major determinants of local tumour control after fractionated irradiation. Radiother. Oncol. 83, 304–310 (2007).
Lambin, P. et al. The ESTRO Breur Lecture 2009. From population to voxel-based radiotherapy: exploiting intra-tumour and intra-organ heterogeneity for advanced treatment of non-small cell lung cancer. Radiother. Oncol. 96, 145–152 (2010).
Prokopiou, S. et al. A proliferation saturation index to predict radiation response and personalize radiotherapy fractionation. Radiat. Oncol. 10, 159 (2015).
Yin, Q. et al. Associations between tumor vascularity, vascular endothelial growth factor expression and PET/MRI radiomic signatures in primary clear-cell-renal-cell-carcinoma: proof-of-concept study. Sci. Rep. 7, 43356 (2017).
Menegakis, A. et al. Residual γH2AX foci after ex vivo irradiation of patient samples with known tumour-type specific differences in radio-responsiveness. Radiother. Oncol. 116, 480–485 (2015).
Menegakis, A. et al. γH2AX assay in ex vivo irradiated tumour specimens: A novel method to determine tumour radiation sensitivity in patient-derived material. Radiother. Oncol. 116, 473–479 (2015).
Slonina, D. & Gasinska, A. Intrinsic radiosensitivity of healthy donors and cancer patients as determined by the lymphocyte micronucleus assay. Int. J. Radiat. Biol. 72, 693–701 (1997).
Fertil, B. & Malaise, E. P. Intrinsic radiosensitivity of human cell lines is correlated with radioresponsiveness of human tumors: analysis of 101 published survival curves. Int. J. Radiat. Oncol. Biol. Phys. 11, 1699–1707 (1985).
Menegakis, A. et al. Prediction of clonogenic cell survival curves based on the number of residual DNA double strand breaks measured by γH2AX staining. Int. J. Radiat. Biol. 85, 1032–1041 (2009).
Bjork-Eriksson, T., West, C., Karlsson, E. & Mercke, C. Tumor radiosensitivity (SF2) is a prognostic factor for local control in head and neck cancers. Int. J. Radiat. Oncol. Biol. Phys. 46, 13–19 (2000).
Chitnis, M. M. et al. IGF-1R inhibition enhances radiosensitivity and delays double-strand break repair by both non-homologous end-joining and homologous recombination. Oncogene 33, 5262–5273 (2014).
Du, S. et al. Attenuation of the DNA damage response by transforming growth factor-beta inhibitors enhances radiation sensitivity of non-small-cell lung cancer cells in vitro and in vivo. Int. J. Radiat. Oncol. Biol. Phys. 91, 91–99 (2015).
Kahn, J. et al. The mTORC1/mTORC2 inhibitor AZD2014 enhances the radiosensitivity of glioblastoma stem-like cells. Neuro Oncol. 16, 29–37 (2014).
West, C. M., Davidson, S. E., Roberts, S. A. & Hunter, R. D. The independence of intrinsic radiosensitivity as a prognostic factor for patient response to radiotherapy of carcinoma of the cervix. Br. J. Cancer 76, 1184–1190 (1997).
Cheng, Q. et al. Development and evaluation of an online three-level proton versus photon decision support prototype for head and neck cancer — Comparison of dose, toxicity and cost-effectiveness. Radiother. Oncol. 118, 281–285 (2016).
Okada, H. et al. Immunotherapy response assessment in neuro-oncology: a report of the RANO working group. Lancet Oncol. 16, e534–e542 (2015).
Tang, C. et al. Pathology-based non-small cell lung cancer radiomics signature describing the local tumor immune environment: discovery and validation. Int. J. Radi. Oncol. Biol. Phys. 96, S42–S43 (2016).
Formenti, S. C. & Demaria, S. Combining radiotherapy and cancer immunotherapy: a paradigm shift. J. Natl Cancer Inst. 105, 256–265 (2013).
Coulie, P. G., Van den Eynde, B. J., van der Bruggen, P. & Boon, T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat. Rev. Cancer 14, 135–146 (2014).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Mellman, I. & Steinman, R. M. Dendritic cells: specialized and regulated antigen processing machines. Cell 106, 255–258 (2001).
Demaria, S., Golden, E. B. & Formenti, S. C. Role of local radiation therapy in cancer immunotherapy. JAMA Oncol. 1, 1325–1332 (2015).
Golden, E. B. et al. Local radiotherapy and granulocyte-macrophage colony-stimulating factor to generate abscopal responses in patients with metastatic solid tumours: a proof-of-principle trial. Lancet Oncol. 16, 795–803 (2015).
Garon, E. B. et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 372, 2018–2028 (2015).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Sanghera, S., Barton, P., Bhattacharya, S., Horne, A. W. & Roberts, T. E. Pharmaceutical treatments to prevent recurrence of endometriosis following surgery: a model-based economic evaluation. BMJ Open 6, e010580 (2016).
Carvalho, S. et al. Early variation of FDG-PET radiomics features in NSCLC is related to overall survival — the “delta radiomics” concept. Radiother. Oncol. 118, S20–S21 (2016).
Fave, X. et al. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med. Phys. 42, 6784–6797 (2015).
Leijenaar, R. T. H. et al. The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis. Sci. Rep. 5, 11075 (2015).
Fave, X. et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci. Rep. 7, 588 (2017).
Deasy, J. O. et al. Improving normal tissue complication probability models: the need to adopt a “data-pooling” culture. Int. J. Radi. Oncol. Biol. Phys. 76, S151–S154 (2010).
Skripcak, T. et al. Creating a data exchange strategy for radiotherapy research: Towards federated databases and anonymised public datasets. Radiother. Oncol. 113, 303–309 (2014).
Budin-Ljosne, I. et al. DataSHIELD: an ethically robust solution to multiple-site individual-level data analysis. Public Health Genomics 18, 87–96 (2015).
Schilsky, R. L., Michels, D. L., Kearbey, A. H., Yu, P. P. & Hudis, C. A. Building a rapid learning health care system for oncology: the regulatory framework of CancerLinQ. J. Clin. Oncol. 32, 2373–2379 (2014).
MAASTRO clinic. euroCAT: Distributed Learning for Individualized Medicine. youtube.com. http://youtu.be/ZDJFOxpwqEA. (2014).
The Cancer Imaging Archive. TCIA Collections. cancerimagingarchive.net http://www.cancerimagingarchive.net/ (2017).
National Cancer Institute, Division of Cancer Treatment & Diagnosis. Quantitative Imaging Network (QIN) [online], (2017).
Radiological Society of North America. Quantitative Imaging Biomarkers Alliance® (QIBA®). rsna.org https://www.rsna.org/qiba/ (2017).
QuiC ConCePT. quic-concept.eu http://www.quic-concept.eu/ (2017).
Benedict, S. H. et al. Overview of the American Society for Radiation Oncology–National Institutes of Health–American Association of Physicists in Medicine Workshop 2015: exploring opportunities for radiation oncology in the era of big data. Int. J. Radi. Oncol. Biol. Phys. 95, 873–879 (2016).
Meldolesi, E. et al. An umbrella protocol for standardized data collection (SDC) in rectal cancer: a prospective uniform naming and procedure convention to support personalized medicine. Radiother. Oncol. 112, 59–62 (2014).
EuroCAT Umbrella Protocol for NSCLC. CancerData.org https://www.cancerdata.org/resource/doi%3A10.17195/candat.2013.08.1 (2017 May 18 th).
van Rossum, P. S. et al. The incremental value of subjective and quantitative assessment of 18F-FDG PET for the prediction of pathologic complete response to preoperative chemoradiotherapy in esophageal cancer. J. Nucl. Med. 57, 691–700 (2016).
Huang, Y. Q. et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 34, 2157–2164 (2016).
Coroller, T. P. et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 114, 345–350 (2015).
Huynh, E. et al. CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother. Oncol. 120, 258–266 (2016).
Cunliffe, A. et al. Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Int. J. Radiat. Oncol. Biol. Phys. 91, 1048–1056 (2015).
Liang, C. et al. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget 7, 31401–31412 (2016).
Hawkins, S. et al. Predicting malignant nodules from screening CT scans. J. Thorac. Oncol. 11, 2120–2128 (2016).
Grossmann, P. et al. Imaging-genomics reveals driving pathways of MRI derived volumetric tumor phenotype features in glioblastoma. BMC Cancer 16, 611 (2016).
Huang, Y. et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 281, 947–957 (2016).
Leijenaar, R. T. et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 54, 1423–1429 (2015).
Cui, Y. et al. Quantitative analysis of 18F-fluorodeoxyglucose positron emission tomography identifies novel prognostic imaging biomarkers in locally advanced pancreatic cancer patients treated with stereotactic body radiation therapy. Int. J. Radiat. Oncol. Biol. Phys. 96, 102–109 (2016).
Li, H. et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology 281, 382–391 (2016).
Acknowledgements
The authors acknowledge financial support from ERC advanced grant (ERC-ADG-2015, no. 694812) and the QuIC-ConCePT project, which is partly funded by EFPI A companies and the Innovative Medicine Initiative Joint Undertaking (IMI JU) under Grant Agreement no. 115151. This research is also supported by the Dutch Technology Foundation STW (grant no. 10696 duCAT & P14-19 Radiomics STRaTegy), which is the applied science division of NWO, and the Technology Programme of the Ministry of Economic Affairs. Authors also acknowledge financial support from the National Institute of Health (NIH-USA U01 CA 143062–01, Radiomics of NSCLC), EU 7 th framework program (EURECA, ARTFORCE – no. 257144, REQUITE – no. 601826), SME phase 2 (EU proposal 673780 – RAIL), the European Program H2020 (BD2Decide – PHC30-689715, ImmunoSABR – no. 733008, PREDICT - ITN no. 766276), Kankeronderzoekfonds Limburg from the Health Foundation Limburg and the Dutch Cancer Society (KWF UM 2011–5020, KWF UM 2009–4454, KWF MAC 2013–6425, KWF MAC 2013–6089) and Alpe d'HuZes-KWF (DESIGN), Center for Translational Molecular Medicine (TraIT), EUROSTARS (SeDI, CloudAtlas, and DART), Interreg V-A Euregio Meuse-Rhine (“Euradiomics”) and Varian Medical Systems (VATE and ROO).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
A.D., leader of the Knowledge Engineering division at MAASTRO, A.J., T.L., J. v- S. and S.W. declare they receive financial support from Varian Medical Systems, a company developing a rapid learning health-care system. R.L. is a salaried employee of, and T.D. consults for ptTheragnostic B.V., a company developing biomarkers and software to individualize radiotherapy treatment. R.T.H.L. and P.L. are co-inventors of radiomics patents (EP2793164A1, US20160203599A1, and WO 2016060557A1).
Supplementary information
Supplementary information
Supplementary Material (PDF 4264 kb)
Glossary
- Phantom studies
-
An artificial structure that imitates human tissue properties is scanned on multiple machines to characterize scan output against a known physical standard.
- Calibration-in-the-large
-
Describes whether the predictions deviate systematically (intercept), whereas the calibration slope should ideally be equal to 1.
- The independence assumption
-
The definition in terms of conditional probabilities is that the probability of B is not changed by knowing that A has occurred. Statistically independent variables are always uncorrelated, but the converse is not necessarily true.
- Feature discretization
-
The process of converting continuous features to discrete binned interval features.
- Bootstrapping
-
Measures the accuracy (defined in terms of bias, variance, confidence intervals, prediction error, etc.) to characterize the sample distribution by way of repeated random sampling methods.
Rights and permissions
About this article
Cite this article
Lambin, P., Leijenaar, R., Deist, T. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14, 749–762 (2017). https://doi.org/10.1038/nrclinonc.2017.141
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2017.141
This article is cited by
-
Standardizing digital biobanks: integrating imaging, genomic, and clinical data for precision medicine
Journal of Translational Medicine (2024)
-
Prediction of malignant esophageal fistula in esophageal cancer using a radiomics-clinical nomogram
European Journal of Medical Research (2024)
-
CT-based radiomics combined with hematologic parameters for survival prediction in locally advanced esophageal cancer patients receiving definitive chemoradiotherapy
Insights into Imaging (2024)
-
A novel sub-regional radiomics model to predict immunotherapy response in non-small cell lung carcinoma
Journal of Translational Medicine (2024)
-
Lymph node metastasis prediction and biological pathway associations underlying DCE-MRI deep learning radiomics in invasive breast cancer
BMC Medical Imaging (2024)