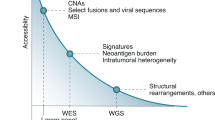
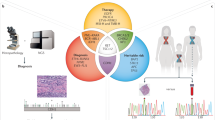
Key Points
-
The landscape of translational oncology has shifted dramatically over the past 10 years, characterized by the introduction of ever-more-sophisticated molecular tools into the clinic
-
Translational cancer-biology studies have markedly improved preclinical models applicable for therapeutics development, as well as our understanding of the roles of inflammation and altered intermediary metabolism in carcinogenesis
-
Translational cancer diagnostics and therapeutics have been revolutionized by the molecular characterization of human tumours, a process that now underlies the development of molecularly-targeted, rather than broadly cytotoxic, anticancer therapies
-
Improvements in molecular tumour-classification techniques will permit their widespread application for patients at diagnosis, disease recurrence, and during therapy, supporting continuous adaptation of therapeutic approaches to evolving tumour characteristics
Abstract
International efforts to sequence the genomes of various human cancers have been broadly deployed in drug discovery programmes. Diagnostic tests that predict the value of the molecularly targeted anticancer agents used in such programmes are conceived and validated in parallel with new small-molecule treatments and immunotherapies. This approach has been aided by better preclinical cancer models; an enhanced appreciation of the complex interactions that exist between tumour cells and their microenvironment; the elucidation of interactions between many of the genetic drivers of cancer, including oncogenes and tumour suppressors; and recent insights into the genetic heterogeneity of human tumours made possible by extraordinary improvements in DNA-sequencing techniques. These advances are being employed in the first generation of genomic clinical trials that will examine the feasibility of matching a broad range of systemic therapies to specific molecular tumour characteristics. More-extensive molecular characterization of tumours and their supporting matrices are anticipated to become standard aspects of oncological practice, permitting continuous molecular re-evaluations of human malignancies on a patient-by-patient and treatment-by-treatment basis. We review selected developments in translational cancer biology, diagnostics, and therapeutics that have occurred over the past decade and offer our thoughts on future prospects for the next few years.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Luo, J., Solimini, N. L. & Elledge, S. J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837 (2009).
Lieu, C. H., Tan, A. C., Leong, S., Diamond, J. R. & Eckhardt, S. G. From bench to bedside: lessons learned in translating preclinical studies in cancer drug development. J. Natl Cancer Inst. 105, 1441–1456 (2013).
Dowell, J. E. & Minna, J. D. The impact of epidermal-growth-factor-receptor mutations in response to lung-cancer therapy. Nat. Clin. Pract. Oncol. 1, 2–3 (2004).
Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Pao, W. et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl Acad. Sci. USA 101, 13306–13311 (2004).
Slamon, D. J. et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 344, 783–792 (2001).
Druker, B. J. Perspectives on the development of imatinib and the future of cancer research. Nat. Med. 15, 1149–1152 (2009).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Hudson, T. J. et al. International network of cancer genome projects. Nature 464, 993–998 (2010).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Garnett, M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570–575 (2012).
Abaan, O. D. et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 73, 4372–4382 (2013).
Kwak, E. L. et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363, 1693–1703 (2010).
Flaherty, K. T. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Fong, P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 (2009).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Hamid, O. et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 369, 134–144 (2013).
Tran, E. et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014).
Palechor-Ceron, N. et al. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. Am. J. Pathol. 183, 1862–1870 (2013).
Perez-Mancera, P. A., Guerra, C., Barbacid, M. & Tuveson, D. A. What we have learned about pancreatic cancer from mouse models. Gastroenterology 142, 1079–1092 (2012).
Siolas, D. & Hannon, G. J. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 73, 5315–5319 (2013).
Hodgkinson, C. L. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014).
Garraway, L. A. & Lander, E. S. Lessons from the cancer genome. Cell 153, 17–37 (2013).
Abrams, J. et al. National Cancer Institute's precision medicine initiatives for the new National Clinical Trials Network. Am. Soc. Clin. Oncol. Educ. Book 34, 71–76 (2014).
Iyer, G. et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 338, 221 (2012).
Sausville, E. A. & Burger, A. M. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 66, 3351–3354 (2006).
Johnson, J. I. et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br. J. Cancer 84, 1424–1431 (2001).
Frese, K. K. & Tuveson, D. A. Maximizing mouse cancer models. Nat. Rev. Cancer 7, 645–658 (2007).
Singh, M., Murriel, C. L. & Johnson, L. Genetically engineered mouse models: closing the gap between preclinical data and trial outcomes. Cancer Res. 72, 2695–2700 (2012).
Navas, C. et al. EGF receptor signaling is essential for K-Ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell 22, 318–330 (2012).
Tentler, J. J. et al. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 9, 338–350 (2012).
Li, S. et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4, 1116–1130 (2013).
Kopetz, S., Lemos, R. & Powis, G. The promise of patient-derived xenografts: the best laid plans of mice and men. Clin. Cancer Res. 18, 5160–5162 (2012).
Suprynowicz, F. A. et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl Acad. Sci. USA 109, 20035–20040 (2012).
Liu, X. et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 180, 599–607 (2012).
Sato, T. & Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013).
Sachs, N. & Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 24, 68–73 (2014).
Trinchieri, G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu. Rev. Immunol. 30, 677–706 (2012).
Guerra, C. et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell 19, 728–739 (2011).
Grivennikov, S. I. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin. Immunopathol. 35, 229–244 (2013).
Wu, Y., Antony, S., Meitzler, J. L. & Doroshow, J. H. Molecular mechanisms underlying chronic inflammation-associated cancers. Cancer Lett. 345, 164–173 (2014).
Thorat, M. A. & Cuzick, J. Role of aspirin in cancer prevention. Curr. Oncol. Rep. 15, 533–540 (2013).
Iida, N. et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970 (2013).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Jones, R. G. & Thompson, C. B. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 23, 537–548 (2009).
Galluzzi, L., Kepp, O., Vander Heiden, M. G. & Kroemer, G. Metabolic targets for cancer therapy. Nat. Rev. Drug Discov. 12, 829–846 (2013).
Vander Heiden, M. G. Exploiting tumor metabolism: challenges for clinical translation. J. Clin. Invest. 123, 3648–3651 (2013).
Kaelin, W. G. Jr & McKnight, S. L. Influence of metabolism on epigenetics and disease. Cell 153, 56–69 (2013).
Rohle, D. et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340, 626–630 (2013).
Wang, F. et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340, 622–626 (2013).
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Chung, C. C. & Chanock, S. J. Current status of genome-wide association studies in cancer. Hum. Genet. 130, 59–78 (2011).
Kim, H. S., Minna, J. D. & White, M. A. GWAS meets TCGA to illuminate mechanisms of cancer predisposition. Cell 152, 387–389 (2013).
Robb, J. A. et al. A call to standardize preanalytic data elements for biospecimens. Arch. Pathol. Lab. Med. 138, 526–537 (2014).
Engel, K. B., Vaught, J. & Moore, H. M. National Cancer Institute biospecimen evidence-based practices: a novel approach to pre-analytical standardization. Biopreserv. Biobank. 12, 148–150 (2014).
Baker, A. F. et al. Stability of phosphoprotein as a biological marker of tumor signaling. Clin. Cancer Res. 11, 4338–4340 (2005).
Kinders, R. J. et al. Preclinical modeling of a phase 0 clinical trial: qualification of a pharmacodynamic assay of poly (ADP-ribose) polymerase in tumor biopsies of mouse xenografts. Clin. Cancer Res. 14, 6877–6885 (2008).
Park, S. R. et al. Validation of a hypoxia-inducible factor-1 alpha specimen collection procedure and quantitative enzyme-linked immunosorbent assay in solid tumor tissues. Anal. Biochem. 459, 1–11 (2014).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Shen, H. & Laird, P. W. Interplay between the cancer genome and epigenome. Cell 153, 38–55 (2013).
Zhang, B. et al. Proteogenomic characterization of human colon and rectal cancer. Nature http://dx.doi.org/10.1038/nature13438.
Callaway, E. Global genomic data-sharing effort kicks off. Nature http://dx.doi.org/10.1038/nature.2014.14826.
Meacham, C. E. & Morrison, S. J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013).
Gerlinger, M. et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 46, 225–233 (2014).
Bedard, P. L., Hansen, A. R., Ratain, M. J. & Siu, L. L. Tumour heterogeneity in the clinic. Nature 501, 355–364 (2013).
Yap, T. A., Gerlinger, M., Futreal, P. A., Pusztai, L. & Swanton, C. Intratumor heterogeneity: seeing the wood for the trees. Sci. Transl. Med. 4, 127ps10 (2012).
Krebs, M. G. et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat. Rev. Clin. Oncol. 11, 129–144 (2014).
Haber, D. A. & Velculescu, V. E. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 4, 650–661 (2014).
Wang, L. H. et al. Monitoring drug-induced γH2AX as a pharmacodynamic biomarker in individual circulating tumor cells. Clin. Cancer Res. 16, 1073–1084 (2010).
Kummar, S., Gutierrez, M., Doroshow, J. H. & Murgo, A. J. Drug development in oncology: classical cytotoxics and molecularly targeted agents. Br. J. Clin. Pharmacol. 62, 15–26 (2006).
Jeong, W., Doroshow, J. H. & Kummar, S. United States Food and Drug Administration approved oral kinase inhibitors for the treatment of malignancies. Curr. Probl. Cancer 37, 110–144 (2013).
Huang, M., Shen, A., Ding, J. & Geng, M. Molecularly targeted cancer therapy: some lessons from the past decade. Trends Pharmacol. Sci. 35, 41–50 (2014).
Kummar, S. et al. Compressing drug development timelines in oncology using phase '0' trials. Nat. Rev. Cancer 7, 131–139 (2007).
Yap, T. A., Sandhu, S. K., Workman, P. & de Bono, J. S. Envisioning the future of early anticancer drug development. Nat. Rev. Cancer 10, 514–523 (2010).
[No authors listed] Pharmacogenomics at work. Nat. Biotechnol. 16, 885 (1998).
Dedes, K. J. et al. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle 10, 1192–1199 (2011).
Kim, E. S. et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 1, 44–53 (2011).
Sequist, L. V. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 3, 75ra26 (2011).
Dias-Santagata, D. et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol. Med. 2, 146–158 (2010).
Roychowdhury, S. et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci. Transl. Med. 3, 111ra121 (2011).
US National Library of Medicine. Clinicaltrials.gov [online], (2014).
Conley, B. A. & Doroshow, J. H. Molecular analysis for therapy choice: NCI MATCH. Semin. Oncol. 41, 297–299 (2014).
Cohen, R. L. & Settleman, J. From cancer genomics to precision oncology—tissue's still an issue. Cell 157, 1509–1514 (2014).
Sarker, D. & Workman, P. Pharmacodynamic biomarkers for molecular cancer therapeutics. Adv. Cancer Res. 96, 213–268 (2007).
Doroshow, J. H. & Parchment, R. E. Oncologic phase 0 trials incorporating clinical pharmacodynamics: from concept to patient. Clin. Cancer Res. 14, 3658–3663 (2008).
Gainor, J. F., Longo, D. L. & Chabner, B. A. Pharmacodynamic biomarkers: falling short of the mark? Clin. Cancer Res. 20, 2587–2594 (2014).
Kinders, R. et al. Implementation of validated pharmacodynamic assays in multiple laboratories: challenges, successes, and limitations. Clin. Cancer Res. 20, 2578–2586 (2014).
Mateo, J., Ong, M., Tan, D. S., Gonzalez, M. A. & de Bono, J. S. Appraising iniparib, the PARP inhibitor that never was—what must we learn? Nat. Rev. Clin. Oncol. 10, 688–696 (2013).
Kinders, R. J. et al. Preclinical modeling of a phase 0 clinical trial: qualification of a pharmacodynamic assay of poly (ADP-ribose) polymerase in tumor biopsies of mouse xenografts. Clin. Cancer Res. 14, 6877–6885 (2008).
Kinders, R. et al. Development of a validated immunofluorescence assay for γH2AX as a pharmacodynamic marker of topoisomerase I inhibitor activity. Clin. Cancer Res. 16, 5447–5457 (2010).
Kummar, S. et al. Phase I study of ABT-888, a PARP inhibitor, in combination with topotecan hydrochloride in adults with refractory solid tumors and lymphomas. Cancer Res. 71, 5626–5634 (2011).
Marrero, A. M. et al. A multiplex quantitative immunofluorescence assay for DNA damage repair in response to cytotoxic treatment [abstract]. Cancer Res. 72 (Suppl. 1), a3620 (2012).
Akbani, R. et al. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report: the RPPA (reverse phase protein array) society. Mol. Cell Proteomics 13, 1625–1643 (2014).
Hayashi, N. et al. Reverse-phase protein array for prediction of patients at low risk of developing bone metastasis from breast cancer. Oncologist http://dx.doi.org/10.1634/theoncologist.2014-0099.
Kelloff, G. J. & Sigman, C. C. Cancer biomarkers: selecting the right drug for the right patient. Nat. Rev. Drug Discov. 11, 201–214 (2012).
Molinari, F. et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin. Cancer Res. 17, 4901–4914 (2011).
Drukker, C. A. et al. Long-term impact of the 70-gene signature on breast cancer outcome. Breast Cancer Res. Treat. 143, 587–592 (2014).
Micheel, C. M., Nass, S. J. & Omenn, G. S. Evolution of tranlational omics: lessons learned and the path forward (The National Academies Press, 2012).
McShane, L. M. et al. Criteria for the use of omics-based predictors in clinical trials: explanation and elaboration. BMC Med. 11, 220 (2013).
Parkinson, D. R. et al. Evidence of clinical utility: an unmet need in molecular diagnostics for patients with cancer. Clin. Cancer Res. 20, 1428–1444 (2014).
Alifrangis, C. C. & McDermott, U. Reading between the lines: understanding drug response in the post genomic era. Mol. Oncol. http://dx.doi.org/10.1016/j.molonc.2014.05.014.
Rebucci, M. & Michiels, C. Molecular aspects of cancer cell resistance to chemotherapy. Biochem. Pharmacol. 85, 1219–1226 (2013).
Park, S. R., Davis-Millin, M., Doroshow, J. H. & Kummar, S. Safety and feasibility of targeted agent combinations in solid tumors. Nat. Rev. Clin. Oncol. 10, 154–168 (2013).
Kummar, S. et al. Utilizing targeted cancer therapeutic agents in combination: novel approaches and urgent requirements. Nat. Rev. Drug Discov. 9, 843–856 (2010).
Holbeck, S., Collins, J. M. & Doroshow, J. H. NCI-60 combination screening matrix of approved anticancer drugs [abstract 27]. Eur. J. Cancer 48 (Suppl. 6), 11 (2012).
Hinrichs, C. S. & Rosenberg, S. A. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol. Rev. 257, 56–71 (2014).
Maus, M. V., Grupp, S. A., Porter, D. L. & June, C. H. Antibody-modified T cells: CARs take the front seat for hematologic malignancies. Blood 123, 2625–2635 (2014).
Dotti, G., Gottschalk, S., Savoldo, B. & Brenner, M. K. Design and development of therapies using chimeric antigen receptor-expressing T cells. Immunol. Rev. 257, 107–126 (2014).
Kantoff, P. W. et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 363, 411–422 (2010).
Mishra, A., Sullivan, L. & Caligiuri, M. A. Molecular pathways: interleukin-15 signaling in health and in cancer. Clin. Cancer Res. 20, 2044–2050 (2014).
Robert, C. et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 364, 2517–2526 (2011).
Fecher, L. A., Agarwala, S. S., Hodi, F. S. & Weber, J. S. Ipilimumab and its toxicities: a multidisciplinary approach. Oncologist 18, 733–743 (2013).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465 (2012).
Schiller, J. T. & Lowy, D. R. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat. Rev. Microbiol. 10, 681–692 (2012).
Schiller, J. T. & Lowy, D. R. Papillomavirus-like particle vaccines. J. Natl Cancer Inst. Monogr. 50–54 (2001).
Lowy, D. R. & Munger, K. Prognostic implications of HPV in oropharyngeal cancer. N. Engl. J. Med. 363, 82–84 (2010).
Uhlman, M. A., Bing, M. T. & Lubaroff, D. M. Prostate cancer vaccines in combination with additional treatment modalities. Immunol. Res. 59, 236–242 (2014).
Woo, S. R. et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 72, 917–927 (2012).
Naidoo, J., Page, D. B. & Wolchok, J. D. Immune checkpoint blockade. Hematol. Oncol. Clin. North Am. 28, 585–600 (2014).
Cheadle, E. J. et al. CAR T cells: driving the road from the laboratory to the clinic. Immunol. Rev. 257, 91–106 (2014).
Wolchok, J. D. et al. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 369, 122–133 (2013).
Ascierto, P. A. et al. Sequencing of BRAF inhibitors and ipilimumab in patients with metastatic melanoma: a possible algorithm for clinical use. J. Transl. Med. 10, 107 (2012).
Histed, S. N. et al. Review of functional/anatomical imaging in oncology. Nucl. Med. Commun. 33, 349–361 (2012).
Stroobants, S. et al. 18FDG-positron emission tomography for the early prediction of response in advanced soft tissue sarcoma treated with imatinib mesylate (Glivec). Eur. J. Cancer 39, 2012–2020 (2003).
Rosen, M. A. & Schnall, M. D. Dynamic contrast-enhanced magnetic resonance imaging for assessing tumor vascularity and vascular effects of targeted therapies in renal cell carcinoma. Clin. Cancer Res. 13, 770s–776s (2007).
Pampaloni, M. H. & Nardo, L. PET/MRI radiotracers beyond 18F-FDG. PET Clin. 9, 345–349 (2014).
Gaykema, S. B. et al. 89Zr-trastuzumab and 89Zr-bevacizumab PET to evaluate the effect of the HSP90 inhibitor NVP-AUY922 in metastatic breast cancer patients. Clin. Cancer Res. 20, 3945–3954 (2014).
Esteban, J. M., Ahn, C., Battifora, H. & Felder, B. Predictive value of estrogen receptors evaluated by quantitative immunohistochemical analysis in breast cancer. Am. J. Clin. Pathol. 102, S9–S12 (1994).
Jordan, V. C. Proven value of translational research with appropriate animal models to advance breast cancer treatment and save lives: the tamoxifen tale. Br. J. Clin. Pharmacol. http://dx.doi.org/10.1111/bcp.12440.
Linden, H. M. et al. Quantitative fluoroestradiol positron emission tomography imaging predicts response to endocrine treatment in breast cancer. J. Clin. Oncol. 24, 2793–2799 (2006).
US National Library of Medicine. Clinicaltrials.gov [online], (2014).
Beumer, J. H. et al. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU). Cancer Chemother. Pharmacol. 62, 363–368 (2008).
Scott, C. L., Mackay, H. J. & Haluska, P. Jr. Patient-derived xenograft models in gynecologic malignancies. Am. Soc. Clin. Oncol. Educ. Book 34, 258–266 (2014).
Lamontanara, A. J., Gencer, E. B., Kuzyk, O. & Hantschel, O. Mechanisms of resistance to BCR–ABL and other kinase inhibitors. Biochim. Biophys. Acta 1834, 1449–1459 (2013).
Doroshow, J. H. Overcoming resistance to targeted anticancer drugs. N. Engl. J. Med. 369, 1852–1853 (2013).
Kummar, S. et al. Phase I trial of Z-endoxifen with estrogen receptor imaging in adults with refractory hormone receptor-positive breast cancer, desmoid tumors, gynecologic tumors, or other hormone receptor-positive solid tumors [abstract 591]. Eur. J. Cancer 48 (Suppl. 6), 181 (2012).
Bhattacharyya, S. et al. Zirconium-89 labeled panitumumab: a potential immuno-PET probe for HER1-expressing carcinomas. Nucl. Med. Biol. 40, 451–457 (2013).
Chu, W. et al. Development of a PET radiotracer for non-invasive imaging of the reactive oxygen species, superoxide, in vivo. Org. Biomol. Chem. 12, 4421–4431 (2014).
Salomonnson, E., Stacer, A. C., Ehrlich, A., Luker, K. E. & Luker, G. D. Imaging CXCL12–CXCR4 signaling in ovarian cancer therapy. PLoS ONE 8, e51500 (2013).
Aerts, H. J. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Acknowledgements
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Author information
Authors and Affiliations
Contributions
Both authors contributed substantially to all stages of the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Doroshow, J., Kummar, S. Translational research in oncology—10 years of progress and future prospects. Nat Rev Clin Oncol 11, 649–662 (2014). https://doi.org/10.1038/nrclinonc.2014.158
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2014.158
This article is cited by
-
Linking insulin like growth factor-1 (IGF-1) rs6214 gene polymorphism and its serum level with risk of colorectal cancer
Beni-Suef University Journal of Basic and Applied Sciences (2022)
-
Severe testing with high-dimensional omics data for enhancing biomedical scientific discovery
npj Systems Biology and Applications (2022)
-
Implementing a clinical cutting-edge and decision-making activity: an ethnographic teamwork approach to a molecular tumorboard
BMC Health Services Research (2020)
-
Tubeimoside I-induced lung cancer cell death and the underlying crosstalk between lysosomes and mitochondria
Cell Death & Disease (2020)
-
The emerging roles of WBP2 oncogene in human cancers
Oncogene (2020)