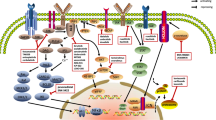
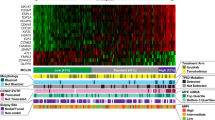
Key Points
-
Lymphomas exhibit heterogeneous genomic alterations, both across and within histological subtypes
-
The molecular heterogeneity underlying lymphoma accounts, at least in part, for variable responses to treatment
-
Lymphoma investigators should incorporate biomarkers into clinical trials to facilitate the clinical development of molecularly targeted therapeutics
-
Techniques for identifying and reporting genomic alterations must be optimized and standardized so that mechanism-based therapies can be effectively applied to the clinical care of lymphoma patients
Abstract
Modern advances in genomics and cancer biology have produced an unprecedented body of knowledge regarding the molecular pathogenesis of lymphoma. The diverse histological subtypes of lymphoma are molecularly heterogeneous, and most likely arise from distinct oncogenic mechanisms. In parallel to these advances in lymphoma biology, several new classes of molecularly targeted agents have been developed with varying degrees of efficacy across the different types of lymphoma. In general, the development of new drugs for treating lymphoma has been mostly empiric, with a limited knowledge of the molecular target, its involvement in the disease, and the effect of the drug on the target. Thus, the variability observed in clinical responses likely results from underlying molecular heterogeneity. In the era of personalized medicine, the challenge for the treatment of patients with lymphoma will involve correctly matching a molecularly targeted therapy to the unique genetic and molecular composition of each individual lymphoma. In this Review, we discuss current and emerging biomarkers that can guide treatment decisions for patients with lymphoma, and explore the potential challenges and strategies for making biomarker-driven personalized medicine a reality in the cure and management of this disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
International Agency for Research on Cancer. Globocan 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012 [online], (2012).
Campo, E. et al. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood 117, 5019–5032 (2011).
Shankland, K. R., Armitage, J. O. & Hancock, B. W. Non-Hodgkin lymphoma. Lancet 380, 848–857 (2012).
Vose, J. et al. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J. Clin. Oncol. 26, 4124–4130 (2008).
Roschewski, M., Staudt, L. M. & Wilson, W. H. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat. Rev. Clin. Oncol. 11, 12–23 (2014).
Borchmann, P., Eichenauer, D. A. & Engert, A. State of the art in the treatment of Hodgkin lymphoma. Nat. Rev. Clin. Oncol. 9, 450–459 (2012).
Ng, A. K., LaCasce, A. & Travis, L. B. Long-term complications of lymphoma and its treatment. J. Clin. Oncol. 29, 1885–1892 (2011).
Younes, A. Beyond chemotherapy: new agents for targeted treatment of lymphoma. Nat. Rev. Clin. Oncol. 8, 85–96 (2011).
Coiffier, B. et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d'Etudes des Lymphomes de l'Adulte. Blood 116, 2040–2045 (2010).
Habermann, T. M. et al. Rituximab-CHOP versus CHOP alone or with maintenance rituximab in older patients with diffuse large B-cell lymphoma. J. Clin. Oncol. 24, 3121–3127 (2006).
Pfreundschuh, M. et al. Six versus eight cycles of bi-weekly CHOP-14 with or without rituximab in elderly patients with aggressive CD20+ B-cell lymphomas: a randomised controlled trial (RICOVER-60). Lancet Oncol. 9, 105–116 (2008).
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000).
Rosenwald, A. et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favourable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J. Exp. Med. 198, 851–862 (2003).
Rosenwald, A. et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 346, 1937–1947 (2002).
Lenz, G. et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 359, 2313–2323 (2008).
Dave, S. S. et al. Molecular diagnosis of Burkitt's lymphoma. N. Engl. J. Med. 354, 2431–2442 (2006).
Dave, S. S. et al. Prediction of survival in follicular lymphoma based on molecular features of tumour-infiltrating immune cells. N. Engl. J. Med. 351, 2159–2169 (2004).
Rosenwald, A. et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell 3, 185–197 (2003).
Ngo, V. N. et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441, 106–110 (2006).
Davis, R. E. et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 463, 88–92 (2010).
Ngo, V. N. et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 470, 115–119 (2011).
Compagno, M. et al. Mutations of multiple genes cause deregulation of NF-kappa B in diffuse large B-cell lymphoma. Nature 459, 717–721 (2009).
Lenz, G. et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319, 1676–1679 (2008).
Wilson, W. H. et al. The Bruton's tyrosine kinase (BTK) inhibitor, ibrutinib (PCI-32765), has preferential activity in the ABC subtype of relapsed/refractory de novo diffuse large B-cell lymphoma (DLBCL): interim results of a multicentre, open-label, phase 2 study [abstract]. Blood 120, a686 (2012).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 (2011).
Lohr, J. G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA 109, 3879–3884 (2012).
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Zhang, J. et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl Acad. Sci. USA 110, 1398–1403 (2013).
Schmitz, R. et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490, 116–120 (2012).
Love, C. et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 44, 1321–1325 (2012).
Richter, J. et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 44, 1316–1320 (2012).
Okosun, J. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46, 176–181 (2014).
Pasqualucci, L. et al. Genetics of follicular lymphoma transformation. Cell Rep. 6, 130–140 (2014).
Bea, S. et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc. Natl Acad. Sci. USA 110, 18250–18255 (2013).
Kridel, R. et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood 119, 1963–1971 (2012).
Rossi, D. et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J. Exp. Med. 209, 1537–1551 (2012).
Kiel, M. J. et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J. Exp. Med. 209, 1553–1565 (2012).
Yoo, H. Y. et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat. Genet. 46, 371–375 (2014).
Palomero, T. et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 46, 166–170 (2014).
Sakata-Yanagimoto, M. et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 46, 171–175 (2014).
Yap, D. B. et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459 (2011).
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-centre origin. Nat. Genet. 42, 181–185 (2010).
Pasqualucci, L. et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471, 189–195 (2011).
Challa-Malladi, M. et al. Combined genetic inactivation of beta2-microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 20, 728–740 (2011).
Ying, C. Y. et al. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat. Immunol. 14, 1084–1092 (2013).
Trinh, D. L. et al. Analysis of FOXO1 mutations in diffuse large B-cell lymphoma. Blood 121, 3666–3674 (2013).
Shaffer, A. L. 3rd, Young, R. M. & Staudt, L. M. Pathogenesis of human B cell lymphomas. Annu. Rev. Immunol. 30, 565–610 (2012).
Tanigaki, K. et al. Notch-RBP-J signalling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3, 443–450 (2002).
Saito, T. et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18, 675–685 (2003).
Meyerson, M., Gabriel, S. & Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696 (2010).
Dewey, F. E. et al. Clinical interpretation and implications of whole-genome sequencing. JAMA 311, 1035–1045 (2014).
Wang, Q. et al. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med. 5, 91 (2013).
Morin, R. D. et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122, 1256–1265 (2013).
Intlekofer, A. M. et al. Profiling genomic alterations of diffuse large B-cell lymphoma (DLBCL) at diagnosis, relapse, and transformation, using a novel clinical diagnostic targeted sequencing platform [abstract]. Blood 122, a1761 (2013).
Chin, L., Hahn, W. C., Getz, G. & Meyerson, M. Making sense of cancer genomic data. Genes Dev. 25, 534–555 (2011).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Pleasance, E. D. et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463, 191–196 (2010).
Pleasance, E. D. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190 (2010).
Stamatoyannopoulos, J. A. et al. Human mutation rate associated with DNA replication timing. Nat. Genet. 41, 393–395 (2009).
Wagle, N. et al. High-throughput detection of actionable genomic alterations in clinical tumour samples by targeted, massively parallel sequencing. Cancer Discov. 2, 82–93 (2012).
Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
Hodges, E. et al. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat. Protoc. 4, 960–974 (2009).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009).
Lipson, D. et al. Identification of actionable genomic alterations in haematologic malignancies by a clinical next generation sequencing-based assay [abstract]. Blood 122, a230 (2013).
Younes, A. & Berry, D. A. From drug discovery to biomarker-driven clinical trials in lymphoma. Nat. Rev. Clin. Oncol. 9, 643–653 (2012).
Chi, P., Allis, C. D. & Wang, G. G. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010).
Bereshchenko, O. R., Gu, W. & Dalla-Favera, R. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 32, 606–613 (2002).
Lane, A. A. & Chabner, B. A. Histone deacetylase inhibitors in cancer therapy. J. Clin. Oncol. 27, 5459–5468 (2009).
Cao, R. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 (2002).
Beguelin, W. et al. EZH2 is required for germinal centre formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013).
Sneeringer, C. J. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumour-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA 107, 20980–20985 (2010).
Knutson, S. K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 8, 890–896 (2012).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Qi, W. et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumour cells proliferation. Proc. Natl Acad. Sci. USA 109, 21360–21365 (2012).
US National Library of Medicine. ClinicalTrials.gov [online], (2014).
US National Library of Medicine. ClinicalTrials.gov [online], (2014).
Cairns, R. A. et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood 119, 1901–1903 (2012).
Losman, J. A. & Kaelin, W. G. Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 (2013).
Ward, P. S. et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 (2010).
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair haematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
Lu, C. et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 (2012).
Wang, F. et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340, 622–626 (2013).
Chen, C. et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 27, 1974–1985 (2013).
Wang, M. L. et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N. Engl. J. Med. 369, 507–516 (2013).
Byrd, J. C. et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 369, 32–42 (2013).
Gopal, A. K. et al. PI3Kdelta inhibition by idelalisib in patients with relapsed indolent lymphoma. N. Engl. J. Med. 370, 1008–1018 (2014).
Rahal, R. et al. Pharmacological and genomic profiling identifies NF-kappaB-targeted treatment strategies for mantle cell lymphoma. Nat. Med. 20, 87–92 (2014).
Senftleben, U. et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signalling pathway. Science 293, 1495–1499 (2001).
Young, R. M. & Staudt, L. M. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat. Rev. Drug Discov. 12, 229–243 (2013).
Landau, D. A. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 (2013).
Gerlinger, M. et al. Intratumour heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Cheson, B. D. & Leonard, J. P. Monoclonal antibody therapy for B-cell non-Hodgkin's lymphoma. N. Engl. J. Med. 359, 613–626 (2008).
Green, T. M. et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J. Clin. Oncol. 30, 3460–3467 (2012).
Johnson, N. A. et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J. Clin. Oncol. 30, 3452–3459 (2012).
Hu, S. et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood 121, 4021–4031 (2013).
Huang, X. et al. Activation of the STAT3 signalling pathway is associated with poor survival in diffuse large B-cell lymphoma treated with R-CHOP. J. Clin. Oncol. 31, 4520–4528 (2013).
Kluk, M. J. et al. Gauging NOTCH1 activation in cancer using immunohistochemistry. PLoS ONE 8, e67306 (2013).
Bose, S., Chandran, S., Mirocha, J. M. & Bose, N. The Akt pathway in human breast cancer: a tissue-array-based analysis. Mod. Pathol. 19, 238–245 (2006).
Espinosa, I. et al. Activation of the NF-kappaB signalling pathway in diffuse large B-cell lymphoma: clinical implications. Histopathology 53, 441–449 (2008).
Mengel, M. et al. Inter-laboratory and inter-observer reproducibility of immunohistochemical assessment of the Ki-67 labelling index in a large multi-centre trial. J. Pathol. 198, 292–299 (2002).
Taylor, C. R. & Levenson, R. M. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment II. Histopathology 49, 411–424 (2006).
Walker, R. A. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment I. Histopathology 49, 406–410 (2006).
Smith, N. R. & Womack, C. A matrix approach to guide IHC-based tissue biomarker development in oncology drug discovery. J. Pathol. 232, 190–198 (2014).
Rikova, K. et al. Global survey of phosphotyrosine signalling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007).
Olsen, J. V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signalling networks. Cell 127, 635–648 (2006).
Rush, J. et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 (2005).
Pasqualucci, L. & Dalla-Favera, R. SnapShot: diffuse large B cell lymphoma. Cancer Cell 25, 132–132.e1 (2014).
Wagle, N. et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 4, 546–553 (2014).
Voss, M. H. et al. Tumour genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin. Cancer Res. 20, 1955–1964 (2014).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Sleijfer, S., Bogaerts, J. & Siu, L. L. Designing transformative clinical trials in the cancer genome era. J. Clin. Oncol. 31, 1834–1841 (2013).
Cheson, B. D. et al. Revised response criteria for malignant lymphoma. J. Clin. Oncol. 25, 579–586 (2007).
Milowsky, M. I. et al. Phase II study of everolimus in metastatic urothelial cancer. BJU Int. 112, 462–470 (2013).
Iyer, G. et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 338, 221 (2012).
Phrma. Medicines in Development for Cancer [online], (Accessed July 2014).
Mathews Griner, L. A. et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc. Natl Acad. Sci. USA 111, 2349–2354 (2014).
Oki, Y. et al. Phase I Study of panobinostat plus everolimus in patients with relapsed or refractory lymphoma. Clin. Cancer Res. 19, 6882–6890 (2013).
Dang, C. et al. Dose-dense doxorubicin and cyclophosphamide followed by weekly paclitaxel with trastuzumab and lapatinib in HER2/neu-overexpressed/amplified breast cancer is not feasible because of excessive diarrhoea. J. Clin. Oncol. 28, 2982–2988 (2010).
Siegel, R., Ma, J., Zou, Z. & Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29 (2014).
Adams, J. M. et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 (1985).
McDonnell, T. J. et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell 57, 79–88 (1989).
McDonnell, T. J. & Korsmeyer, S. J. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14; 18). Nature 349, 254–256 (1991).
Strasser, A., Harris, A. W., Bath, M. L. & Cory, S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348, 331–333 (1990).
Fanidi, A., Harrington, E. A. & Evan, G. I. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature 359, 554–556 (1992).
Bissonnette, R. P., Echeverri, F., Mahboubi, A. & Green, D. R. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature 359, 552–554 (1992).
Vandenberg, C. J. & Cory, S. ABT-199, a new Bcl-2-specific BH3 mimetic, has in vivo efficacy against aggressive Myc-driven mouse lymphomas without provoking thrombocytopenia. Blood 121, 2285–2288 (2013).
Souers, A. J. et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumour activity while sparing platelets. Nat. Med. 19, 202–208 (2013).
Sander, S. et al. Synergy between PI3K signalling and MYC in Burkitt lymphomagenesis. Cancer Cell 22, 167–179 (2012).
Kaelin, W. G. Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 5, 689–698 (2005).
Yang, Y. et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 21, 723–737 (2012).
Acknowledgements
A.M.I. would like to thank the Conquer Cancer Foundation of ASCO for funding.
Author information
Authors and Affiliations
Contributions
A.M.I. and A.Y. researched data for article, reviewed and edited the manuscript before submission, substantially contributed to discussion of content and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.M.I. and A.Y. have served as consultants for Foundation Medicine
Rights and permissions
About this article
Cite this article
Intlekofer, A., Younes, A. Precision therapy for lymphoma—current state and future directions. Nat Rev Clin Oncol 11, 585–596 (2014). https://doi.org/10.1038/nrclinonc.2014.137
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2014.137
This article is cited by
-
MLMDA: a machine learning approach to predict and validate MicroRNA–disease associations by integrating of heterogenous information sources
Journal of Translational Medicine (2019)
-
A selective c-Met and Trks inhibitor Indo5 suppresses hepatocellular carcinoma growth
Journal of Experimental & Clinical Cancer Research (2019)
-
The use of PanDrugs to prioritize anticancer drug treatments in a case of T-ALL based on individual genomic data
BMC Cancer (2019)
-
Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets
Leukemia (2018)
-
Integrated DNA/RNA targeted genomic profiling of diffuse large B-cell lymphoma using a clinical assay
Blood Cancer Journal (2018)