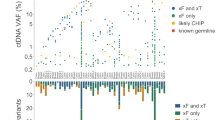
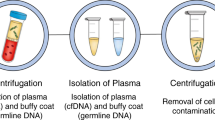
Abstract
The majority of samples in existing tumour biobanks are surgical specimens of primary tumours. Insights into tumour biology, such as intratumoural heterogeneity, tumour–host crosstalk, and the evolution of the disease during therapy, require biospecimens from the primary tumour and those that reflect the patient's disease in specific contexts. Next-generation 'omics' technologies facilitate deep interrogation of tumours, but the characteristics of the samples can determine the ultimate accuracy of the results. The challenge is to biopsy tumours, in some cases serially over time, ensuring that the samples are representative, viable, and adequate both in quantity and quality for subsequent molecular applications. The collection of next-generation biospecimens, tumours, and blood samples at defined time points during the disease trajectory—either for discovery research or to guide clinical decisions—presents additional challenges and opportunities. From an organizational perspective, it also requires new additions to the multidisciplinary therapeutic team, notably interventional radiologists, molecular pathologists, and bioinformaticians. In this Review, we describe the existing procedures for sample procurement and processing of next-generation biospecimens, and highlight the issues involved in this endeavour, including the ethical, logistical, scientific, informational, and financial challenges accompanying next-generation biobanking.
Key Points
-
Next-generation biospecimens are biopsy-type clinical specimens collected from patients at distinct time points and in a prespecified clinical context of treatment, made available for multidimensional high-throughput technologies
-
Biopsies of recurrent primary or metastatic tumours are highly sought after next-generation biospecimens for both research purposes and the clinical management of patients
-
Controlling preanalytical variables is critical to ensure that the results of multidimensional high-throughput profiling are accurate and reproducible
-
Standard operating procedures for biospecimen collection and processing, with quality assurance of every specimen, must be developed and adhered to, with particular emphasis placed on the training of personnel
-
Collection of next-generation biospecimens requires increased resources and a multidisciplinary team consisting of interventional radiologists and molecular pathologists
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
24 September 2013
In the original published version of this article, the link in reference 106 was incorrect and should have referred to the Biospecimen Research Database of the NCI (page 444 of the article). This reference has now been corrected for the HTML and PDF versions of the article.
References
Lovly, C. M. et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS ONE 7, e35309 (2012).
Von Hoff, D. D. et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J. Clin. Oncol. 28, 4877–4883 (2010).
Roychowdhury, S. et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci. Transl. Med. 3, 111ra121 (2011).
Desai, A. N. & Jere, A. Next-generation sequencing: ready for the clinics? Clin. Genet. 81, 503–510 (2012).
Blow, N. Biobanking: freezer burn. Nat. Methods 6, 173–178 (2009).
International Cancer Genome Consortium et al. International network of cancer genome projects. Nature 464, 993–998 (2010).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Cancer Genome Atlas Research Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
The Cancer Genome Atlas. TCGA Tissue Sample Requirements: High Quality Requirements Yield High Quality Data [online], (2013).
Koh, S. S. et al. Differential gene expression profiling of primary cutaneous melanoma and sentinel lymph node metastases. Mod. Pathol. 25, 828–837 (2012).
Shah, S. P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Turajlic, S. et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 22, 196–207 (2012).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Aurilio, G. et al. Discordant hormone receptor and human epidermal growth factor receptor 2 status in bone metastases compared to primary breast cancer. Acta Oncol. http://dx.doi.org/10.3109/0284186X.2012.754990.
Botteri, E. et al. Biopsy of liver metastasis for women with breast cancer: impact on survival. Breast 21, 284–288 (2012).
Gray, J. Cancer: Genomics of metastasis. Nature 464, 989–990 (2010).
Shi, H. et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat. Commun. 3, 724 (2012).
Doebele, R. C. et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin. Cancer Res. 18, 1472–1482 (2012).
Flaherty, K. T. et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med. 367, 1694–1703 (2012).
Gorges, T. M. & Pantel, K. Circulating tumor cells as therapy-related biomarkers in cancer patients. Cancer Immunol. Immunother. 62, 931–939 (2013).
US National Library of Medicine. Clinicaltrials.gov [online], (2013).
Dawson, S. J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Misale, S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486, 532–536 (2012).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Overman, M. J. et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J. Clin. Oncol. 31, 17–22 (2013).
Goulart, B. H. et al. Trends in the use and role of biomarkers in phase I oncology trials. Clin. Cancer Res. 13, 6719–6726 (2007).
Subramanian, J. & Simon, R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J. Natl Cancer Inst. 102, 464–474 (2010).
Liu, G. et al. Pharmacogenetic analysis of BR.21, a placebo-controlled randomized phase III clinical trial of erlotinib in advanced non-small cell lung cancer. J. Thorac. Oncol. 7, 316–322 (2012).
Choi, Y. L. et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 363, 1734–1739 (2010).
Sharma, M. R. & Schilsky, R. L. Role of randomized phase III trials in an era of effective targeted therapies. Nat. Rev. Clin. Oncol. 9, 208–214 (2011).
Amir, E. et al. Tissue confirmation of disease recurrence in breast cancer patients: pooled analysis of multi-centre, multi-disciplinary prospective studies. Cancer Treat. Rev. 38, 708–714 (2011).
Amir, E. et al. Prospective study evaluating the impact of tissue confirmation of metastatic disease in patients with breast cancer. J. Clin. Oncol. 30, 587–592 (2012).
Chia, S. Testing for discordance at metastatic relapse: does it matter? J. Clin. Oncol. 30, 575–576 (2012).
Niikura, N. et al. Loss of human epidermal growth factor receptor 2 (HER2) expression in metastatic sites of HER2-overexpressing primary breast tumors. J. Clin. Oncol. 30, 593–599 (2012).
Watanabe, T. et al. Heterogeneity of KRAS status may explain the subset of discordant KRAS status between primary and metastatic colorectal cancer. Dis. Colon Rectum 54, 1170–1178 (2011).
Artale, S. et al. Mutations of KRAS and BRAF in primary and matched metastatic sites of colorectal cancer. J. Clin. Oncol. 26, 4217–4219 (2008).
Kim, E. et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 1, 44–53 (2011).
Tran, B. et al. Feasibility of real time next generation sequencing of cancer genes linked to drug response: results from a clinical trial. Int. J. Cancer 132, 1547–1555 (2013).
Dancey, J. E., Bedard, P. L., Onetto, N. & Hudson, T. J. The genetic basis for cancer treatment decisions. Cell 148, 409–420 (2012).
Institute of Cancerology Gustave Roussy. Official launch of the clinical, academic and international trial WINTHER : a bioinformatics scoring system that predicts the response to known treatments for each patient [online], (2012).
Tsimberidou, A. M. et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center Initiative. Clin. Cancer Res. 18, 6373–6383 (2012).
Wright, J. R. et al. Why cancer patients enter randomized clinical trials: exploring the factors that influence their decision. J. Clin. Oncol. 22, 4312–4318 (2004).
Peppercorn, J. et al. Ethics of mandatory research biopsy for correlative end points within clinical trials in oncology. J. Clin. Oncol. 28, 2635–2640 (2010).
Lee, J. M. et al. Feasibility and safety of sequential research-related tumor core biopsies in clinical trials. Cancer 119, 1357–1364 (2013).
Olson, E. M., Lin, N. U., Krop, I. E. & Winer, E. P. The ethical use of mandatory research biopsies. Nat. Rev. Clin. Oncol. 8, 620–625 (2011).
Tam, A. L. et al. Feasibility of image-guided transthoracic core-needle biopsy in the BATTLE lung trial. J. Thorac. Oncol. 8, 436–442 (2013).
Dowlati, A. et al. Sequential tumor biopsies in early phase clinical trials of anticancer agents for pharmacodynamic evaluation. Clin. Cancer Res. 7, 2971–2976 (2001).
Nazarian, L. N. et al. Safety and efficacy of sonographically guided random core biopsy for diffuse liver disease. J. Ultrasound Med. 19, 537–541 (2000).
Caliskan, K. C., Cakmakci, E., Celebi, I. & Basak, M. The importance of experience in percutaneous liver biopsies guided with ultrasonography: a lesion-focused approach. Acad. Radiol. 19, 256–259 (2012).
Grant, A. & Neuberger, J. Guidelines on the use of liver biopsy in clinical practice. British Society of Gastroenterology. Gut 45 (Suppl. 4), IV1–IV11 (1999).
Robertson, E. G. & Baxter, G. Tumour seeding following percutaneous needle biopsy: the real story! Clin. Radiol. 66, 1007–1014 (2011).
Agulnik, M., Oza, A. M., Pond, G. R. & Siu, L. L. Impact and perceptions of mandatory tumor biopsies for correlative studies in clinical trials of novel anticancer agents. J. Clin. Oncol. 24, 4801–4807 (2006).
Wolf, S. M. et al. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genet. Med. 14, 361–384 (2012).
Bredenoord, A. L., Kroes, H. Y., Cuppen, E., Parker, M. & van Delden, J. J. Disclosure of individual genetic data to research participants: the debate reconsidered. Trends Genet. 27, 41–47 (2011).
Gymrek, M., McGuire, A. L., Golan, D., Halperin, E. & Erlich, Y. Identifying personal genomes by surname inference. Science 339, 321–324 (2013).
Aguilar-Mahecha, A. et al. Making personalized medicine a reality: the challenges of a modern translational research biopsy-driven program in an academic setting: the Segal Cancer Center experience. J. Med. Person. 9, 104–111 (2011).
Ricci, D. S. et al. Global requirements for DNA sample collections: results of a survey of 204 ethics committees in 40 countries. Clin. Pharmacol. Ther. 89, 554–561 (2011).
Katona, T. M. et al. Genetically heterogeneous and clonally unrelated metastases may arise in patients with cutaneous melanoma. Am. J. Surg. Pathol. 31, 1029–1037 (2007).
Liegl, B. et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J. Pathol. 216, 64–74 (2008).
Taniguchi, K., Okami, J., Kodama, K., Higashiyama, M. & Kato, K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 99, 929–935 (2008).
Yancovitz, M. et al. Intra- and inter-tumor heterogeneity of BRAF(V600E) mutations in primary and metastatic melanoma. PLoS ONE 7, e29336 (2012).
Maley, C. C. et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 38, 468–473 (2006).
Mehra, R. et al. Characterization of bone metastases from rapid autopsies of prostate cancer patients. Clin. Cancer Res. 17, 3924–3932 (2011).
Aguilar-Mahecha, A. et al. Study of pre-analytical variables in plasma and breast biopsies to be used for proteomic and genomic studies [online], Biorepositories and Biospecimen Research Branch http://biospecimens.cancer.gov/meeting/brnsymposium/2011/Posters/Aguilar-508.pdf (2011).
Diaz, Z. et al. Next-generation biobanking of metastases to enable multidimensional molecular profiling in personalized medicine. Mod. Pathol. http://dx.doi.org/10.1038/modpathol.2013.81.
Pusztai, L. et al. Gene expression profiles obtained from fine-needle aspirations of breast cancer reliably identify routine prognostic markers and reveal large-scale molecular differences between estrogen-negative and estrogen-positive tumors. Clin. Cancer Res. 9, 2406–2415 (2003).
Willems, S. M., van Deurzen, C. H. & van Diest, P. J. Diagnosis of breast lesions: fine-needle aspiration cytology or core needle biopsy? A review. J. Clin. Pathol. 65, 287–292 (2012).
von Renteln, D. et al. A novel flexible cryoprobe for EUS-guided pancreatic biopsies. Gastrointest. Endosc. 77, 784–792 (2013).
Hetzel, J. et al. Cryobiopsy increases the diagnostic yield of endobronchial biopsy: a multicentre trial. Eur. Respir. J. 39, 685–690 (2012).
Schumann, C. et al. Cryoprobe biopsy increases the diagnostic yield in endobronchial tumor lesions. J. Thorac. Cardiovasc. Surg. 140, 417–421 (2010).
Masuda, N., Ohnishi, T., Kawamoto, S., Monden, M. & Okubo, K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 27, 4436–4443 (1999).
Srinivasan, M., Sedmak, D. & Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 161, 1961–1971 (2002).
Klockenbusch, C., O'Hara, J. E. & Kast, J. Advancing formaldehyde cross-linking towards quantitative proteomic applications. Anal. Bioanal. Chem. 404, 1057–1067 (2012).
Reis, P. P. et al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 11, 46 (2011).
Malkov, V. A. et al. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res. Notes 2, 80 (2009).
Holley, T. et al. Deep clonal profiling of formalin fixed paraffin embedded clinical samples. PLoS ONE 7, e50586 (2012).
Tuononen, K. et al. Comparison of targeted next-generation sequencing (NGS) and real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on formalin-fixed, paraffin-embedded tumor material of non-small cell lung carcinoma-superiority of NGS. Genes Chromosomes Cancer 52, 503–511 (2013).
Florell, S. R. et al. Preservation of RNA for functional genomic studies: a multidisciplinary tumor bank protocol. Mod. Pathol. 14, 116–128 (2001).
Hatzis, C. et al. Effects of tissue handling on RNA integrity and microarray measurements from resected breast cancers. J. Natl Cancer Inst. 103, 1871–1883 (2011).
Chowdary, D. et al. Prognostic gene expression signatures can be measured in tissues collected in RNAlater preservative. J. Mol. Diagn. 8, 31–39 (2006).
Przybytkowski, E., Aguilar-Mahecha, A., Nabavi, S., Tonellato, P. J. & Basik, M. Ultradense array CGH and discovery of micro-copy number alterations and gene fusions in the cancer genome. Methods Mol. Biol. 973, 15–38 (2013).
Belloni, B. et al. Will PAXgene substitute formalin? A morphological and molecular comparative study using a new fixative system. J. Clin. Pathol. 66, 124–135 (2013).
Kap, M. et al. Histological assessment of PAXgene tissue fixation and stabilization reagents. PLoS ONE 6, e27704 (2011).
Groelz, D. et al. Non-formalin fixative versus formalin-fixed tissue: a comparison of histology and RNA quality. Exp. Mol. Pathol. 94, 188–194 (2013).
Botling, J. & Micke, P. Biobanking of fresh frozen tissue from clinical surgical specimens: transport logistics, sample selection, and histologic characterization. Methods Mol. Biol. 675, 299–306 (2011).
Ellis, M. et al. Development and validation of a method for using breast core needle biopsies for gene expression microarray analyses. Clin. Cancer Res. 8, 1155–1166 (2002).
Ono, M. et al. Tumor-infiltrating lymphocytes are correlated with response to neoadjuvant chemotherapy in triple-negative breast cancer. Breast Cancer Res. Treat. 132, 793–805 (2012).
Loi, S. et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98 J. Clin. Oncol. 31, 860–867 (2013).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Biankin, A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Esgueva, R. et al. Next-generation prostate cancer biobanking: toward a processing protocol amenable for the International Cancer Genome Consortium. Diagn. Mol. Pathol. 21, 61–68 (2012).
Golubeva, Y., Salcedo, R., Mueller, C., Liotta, L. A. & Espina, V. Laser capture microdissection for protein and NanoString RNA analysis. Methods Mol. Biol. 931, 213–257 (2013).
Yuan, Y. et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci. Transl. Med. 4, 157ra143 (2012).
Ki, D. H. et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int. J. Cancer 121, 2005–2012 (2007).
Türeci, O. et al. Computational dissection of tissue contamination for identification of colon cancer-specific expression profiles. FASEB J. 17, 376–385 (2003).
Kotorashvili, A. et al. Effective DNA/RNA co-extraction for analysis of microRNAs, mRNAs, and genomic DNA from formalin-fixed paraffin-embedded specimens. PLoS ONE 7, e34683 (2012).
Soares, A. R., Pereira, P. M. & Santos, M. A. Next-generation sequencing of miRNAs with Roche 454 GS-FLX technology: steps for a successful application. Methods Mol. Biol. 822, 189–204 (2012).
Moore, H. M., Compton, C. C., Alper, J. & Vaught, J. B. International approaches to advancing biospecimen science. Cancer Epidemiol. Biomarkers Prev. 20, 729–732 (2011).
Pazzagli, M. et al. SPIDIA-RNA: first external quality assessment for the pre-analytical phase of blood samples used for RNA based analyses. Methods 59, 20–31 (2013).
Moore, H. M. The NCI Biospecimen Research Network. Biotech. Histochem. 87, 18–23 (2012).
National Cancer Institute. Biospecimen Research Database [online], (2013).
Nature Publishing Group. About protocol exchange. Protocol Exchange [online], (2013).
Aguilar-Mahecha, A., Kuzyk, M. A., Domanski, D., Borchers, C. H. & Basik, M. The effect of pre-analytical variability on the measurement of MRM-MS-based mid- to high-abundance plasma protein biomarkers and a panel of cytokines. PLoS ONE 7, e38290 (2012).
Begley, C. G. & Ellis, L. M. Drug development: Raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
Simeon-Dubach, D., Burt, A. D. & Hall, P. A. Quality really matters: the need to improve specimen quality in biomedical research. J. Pathol. http://dx.doi.org/10.1002/path.4117 (2012).
Ransohoff, D. F. & Gourlay, M. L. Sources of bias in specimens for research about molecular markers for cancer. J. Clin. Oncol. 28, 698–704 (2010).
Taube, S. E. et al. A perspective on challenges and issues in biomarker development and drug and biomarker codevelopment. J. Natl Cancer Inst. 101, 1453–1463 (2009).
Silberman, S. Libraries of flesh: the sorry state of human tissue storage. WIRED Magazine (24 May 2010).
Massett, H. A. et al. Assessing the need for a standardized cancer HUman Biobank (caHUB): findings from a national survey with cancer researchers. J. Natl Cancer Inst. Monogr. 2011, 8–15 (2011).
Betsou, F. et al. Standard preanalytical coding for biospecimens: defining the sample PREanalytical code. Cancer Epidemiol. Biomarkers Prev. 19, 1004–1011 (2010).
Moore, H. M. et al. Biospecimen reporting for improved study quality (BRISQ). Cancer Cytopathol. 119, 92–101 (2011).
Strand, C., Enell, J., Hedenfalk, I. & Fernö, M. RNA quality in frozen breast cancer samples and the influence on gene expression analysis--a comparison of three evaluation methods using microcapillary electrophoresis traces. BMC Mol. Biol. 8, 38 (2007).
Kennedy, R. D. et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J. Clin. Oncol. 29, 4620–4626 (2011).
Georgiou, C. D., Papapostolou, I. & Grintzalis, K. Protocol for the quantitative assessment of DNA concentration and damage (fragmentation and nicks). Nat. Protoc. 4, 125–131 (2009).
Esserman, L. J. et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657). Breast Cancer Res. Treat. 132, 1049–1062 (2012).
Nadkarni, P. M., Kemp, R. & Parikh, C. R. Leveraging a clinical research information system to assist biospecimen data and workflow management: a hybrid approach. J. Clin. Bioinforma 1, 22 (2011).
Welinder, C. et al. Establishing a Southern Swedish Malignant Melanoma OMICS and biobank clinical capability. Clin. Transl. Med. 2, 7 (2013).
National Cancer Informatics Program. NCIP Launch Meeting Summary Report (05/31/2012). National Cancer Informatics Program Launch Meeting [online], (2012).
UC Santa Cruz. UCSC Cancer Genomics Browser [online], (2013).
Goldman, M. et al. The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Res. 41, D949–D954 (2013).
Schroeder, M. P., Gonzalez-Perez, A. & Lopez-Bigas, N. Visualizing multidimensional cancer genomics data. Genome Med. 5, 9 (2013).
Gomez-Roca, C. A. et al. Sequential research-related biopsies in phase I trials: acceptance, feasibility and safety. Ann. Oncol. 23, 1301–1306 (2012).
Baker, M. Biorepositories: Building better biobanks. Nature 486, 141–146 (2012).
Matzke, E. A. M. et al. Certification for biobanks: the program developed by the Canadian Tumor Repository Network (CTRNet). Biopreserv. Biobank. 10, 426–432 (2012).
Harris, J. R. et al. Toward a roadmap in global biobanking for health. Eur. J. Hum. Genet. 20, 1105–1111 (2012).
Watson, R. W., Kay, E. W. & Smith, D. Integrating biobanks: addressing the practical and ethical issues to deliver a valuable tool for cancer research. Nat. Rev. Cancer 10, 646–651 (2010).
Fenstermacher, D. A., Wenham, R. M., Rollison, D. E. & Dalton, W. S. Implementing personalized medicine in a cancer center. Cancer J. 17, 528–536 (2011).
Yachida, S. & Iacobuzio-Donahue, C. A. Evolution and dynamics of pancreatic cancer progression. Oncogene http://dx.doi.org/10.1038/onc.2013.29 (2013).
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012).
Giordano, A. & Cristofanilli, M. CTCs in metastatic breast cancer. Recent Results Cancer Res. 195, 193–201 (2012).
Jacot, W. et al. Lack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profiles. Breast Cancer Res. 13, R133 (2011).
Rodig, S. J. et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin. Cancer Res. 15, 5216–5223 (2009).
Calvo, E. & Baselga, J. Ethnic differences in response to epidermal growth factor receptor tyrosine kinase inhibitors. J. Clin. Oncol. 24, 2158–2163 (2006).
Mudur, G. Indian scientists object to export of human biological material for research. BMJ 325, 990 (2002).
Barker, A. D. et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin. Pharmacol. Ther. 86, 97–100 (2009).
Acknowledgements
The authors would like to recognize the Interventional Radiology Department at the Jewish General Hospital (especially Andre Constantin and Errol Camlioglu), the Pathology Group from Hôpital du Saint-Sacrement (especially Benoit Têtu and Michèle Orain) and the Jewish General Hospital (Adrian Gologan and Tina Haliotis) and the laboratory of Koren Mann, for contributing their expertise to the collection and processing of high-quality biospecimens. The authors would like to thank both Thérèse Gagnon-Kugler and Suzan McNamara of the Québec Clinical Research Organization in Cancer for their valuable comments in drafting the manuscript. We would like to acknowledge support from the FRQS-Réseau de Recherche sur le Cancer, Genome Québec and the Québec Breast Cancer Foundation.
Author information
Authors and Affiliations
Contributions
A. Aguila-Mahecha, M. Basik, G. Batist, Z. Diaz and C. Rousseau researched data and wrote the manuscript. C. M. T. Greenwood, A. Spatz and S. Tejpar made substantial contribution to discussion of content. All authors reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Basik, M., Aguilar-Mahecha, A., Rousseau, C. et al. Biopsies: next-generation biospecimens for tailoring therapy. Nat Rev Clin Oncol 10, 437–450 (2013). https://doi.org/10.1038/nrclinonc.2013.101
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2013.101
This article is cited by
-
Ultrasound-guided biopsy of challenging abdominopelvic targets
Abdominal Radiology (2022)
-
Bleeding management in computed tomography-guided liver biopsies by biopsy tract plugging with gelatin sponge slurry
Scientific Reports (2021)
-
Basic principles of biobanking: from biological samples to precision medicine for patients
Virchows Archiv (2021)
-
Yield and Integrity of RNA from Brain Samples are Largely Unaffected by Pre-analytical Procedures
Neurochemical Research (2021)
-
Circulating tumor DNA and liquid biopsy in oncology
Nature Cancer (2020)