Abstract
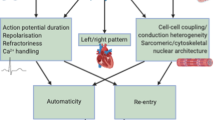
Atrial fibrillation (AF) is the most-common sustained arrhythmia observed in clinical practice, but response to therapy is highly variable between patients. Current drug therapies to suppress AF are incompletely and unpredictably effective and carry substantial risk of proarrhythmia and noncardiac toxicities. The limited success of therapy for AF is partially the result of heterogeneity of the underlying substrate, interindividual differences in disease mechanisms, and our inability to predict response to therapies in individual patients. In this Review, we discuss the evidence that variability in response to drug therapy is also conditioned by the underlying genetic substrate for AF. Increased susceptibility to AF is mediated through diverse genetic mechanisms, including modulation of the atrial action-potential duration, conduction slowing, and impaired cell-to-cell communication, as well as novel mechanisms, such as regulation of signalling proteins important in the pathogenesis of AF. However, the translation of genetic data to the care of the patients with AF has been limited because of poor understanding of the underlying mechanisms associated with common AF-susceptibility loci, a dearth of prospective, adequately powered studies, and the challenges associated with determining efficacy of antiarrhythmic drugs. What is apparent, however, is the need for appropriately designed, genotype-directed clinical trials.
Key Points
-
The incidence of atrial fibrillation (AF) is rising, with approximately 16 million of the US population projected to develop AF by 2050
-
Limitations of current therapies for AF have spurred research into understanding the genetic basis of this arrhythmia
-
Positional cloning and candidate-gene approaches have linked rare genetic variants in ion channels, gap-junction proteins, and signalling molecules with the development of AF
-
Genome-wide association studies have identified nine commonly occurring AF-susceptibility loci associated with cardiopulmonary development, cardiac ion channels, and cell-signalling molecules that might be important in the pathogenesis of AF
-
Both common and rare genetic variants have provided insights into AF genetic mechanisms, but the direct impact of this understanding on the management of patients has been limited
-
Exploiting the genetic mechanisms of AF to prescribe personalized therapy is an important goal, but clinical trials are needed to determine whether genotype-directed treatment of AF is viable
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lloyd-Jones, D. M. et al. Lifetime risk for development of atrial fibrillation: the Framingham Heart Study. Circulation 110, 1042–1046 (2004).
Krahn, A. D., Manfreda, J., Tate, R. B., Mathewson, F. A. & Cuddy, T. E. The natural history of atrial fibrillation: incidence, risk factors, and prognosis in the Manitoba Follow-Up Study. Am. J. Med. 98, 476–484 (1995).
Benjamin, E. J. et al. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation 98, 946–952 (1998).
Darbar, D. et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J. Am. Coll. Cardiol. 41, 2185–2192 (2003).
Piccini, J. P. et al. Pharmacotherapy in Medicare beneficiaries with atrial fibrillation. Heart Rhythm 9, 1403–1408 (2012).
Wyse, D. G. et al. A comparison of rate control and rhythm control in patients with atrial fibrillation. N. Engl. J. Med. 347, 1825–1833 (2002).
Van Gelder, I. C. et al. A comparison of rate control and rhythm control in patients with recurrent persistent atrial fibrillation. N. Engl. J. Med. 347, 1834–1840 (2002).
Opolski, G. et al. Rate control vs rhythm control in patients with nonvalvular persistent atrial fibrillation: the results of the Polish How to Treat Chronic Atrial Fibrillation (HOT CAFE) study. Chest 126, 476–486 (2004).
Corley, S. D. et al. Relationships between sinus rhythm, treatment, and survival in the Atrial Fibrillation Follow-Up Investigation of Rhythm Management (AFFIRM) study. Circulation 109, 1509–1513 (2004).
Fuster, V. et al. ACC/AHA/ESC 2006 guidelines for the management of patients with atrial fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to revise the 2001 guidelines for the management of patients with atrial fibrillation): developed in collaboration with the European Heart Rhythm Association and the Heart Rhythm Society. Circulation 114, e257–e354 (2006).
Benjamin, E. J. et al. Prevention of atrial fibrillation: report from a national Heart, Lung, and Blood Institute workshop. Circulation 119, 606–618 (2009).
Darbar, D. Genetics of atrial fibrillation: rare mutations, common polymorphisms, and clinical relevance. Heart Rhythm 5, 483–486 (2008).
Wolff, L. Familial auricular fibrillation. N. Engl. J. Med. 229, 396–398 (1943).
Ellinor, P. T., Yoerger, D. M., Ruskin, J. N. & Macrae, C. A. Familial aggregation in lone atrial fibrillation. Hum. Genet. 118, 179–184 (2005).
Arnar, D. O. et al. Familial aggregation of atrial fibrillation in Iceland. Eur. Heart J. 27, 708–712 (2006).
Christophersen, I. E. et al. Familial aggregation of atrial fibrillation: a study in Danish twins. Circ. Arrhythm. Electrophysiol. 2, 378–383 (2009).
Fox, C. S. et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA 291, 2851–2855 (2004).
Marcus, G. M. et al. A first-degree family history in lone atrial fibrillation patients. Heart Rhythm 5, 826–830 (2008).
Lubitz, S. A. et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA 304, 2263–2269 (2010).
Brugada, R. et al. Identification of a genetic locus for familial atrial fibrillation. N. Engl. J. Med. 336, 905–911 (1997).
Ellinor, P. T., Shin, J. T., Moore, R. K., Yoerger, D. M. & MacRae, C. A. Locus for atrial fibrillation maps to chromosome 6q14-16. Circulation 107, 2880–2883 (2003).
Schott, J. J., Probst, V. & Mabo, P. A new locus for atrial fibrillation maps to chromosome 20q12-13. Circulation 110, 1245 (2004).
Oberti, C. et al. Genome-wide linkage scan identifies a novel genetic locus on chromosome 5p13 for neonatal atrial fibrillation associated with sudden death and variable cardiomyopathy. Circulation 110, 3753–3759 (2004).
Volders, P. G. et al. Mapping a novel locus for familial atrial fibrillation on chromosome 10p11-q21. Heart Rhythm 4, 469–475 (2007).
Chen, Y. H. et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science 299, 251–254 (2003).
Hodgson-Zingman, D. M. et al. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N. Engl. J. Med. 359, 158–165 (2008).
Mann, S. A. et al. Epistatic effects of potassium channel variation on cardiac repolarization and atrial fibrillation risk. J. Am. Coll. Cardiol. 59, 1017–1025 (2012).
Darbar, D., Parvez, B. & Abraham, R. Repolarization recipes for atrial fibrillation: beyond single channel variants. J. Am. Coll. Cardiol. 59, 1026–1028 (2012).
Ritchie, M. D. et al. Chromosome 4q25 variants are genetic modifiers of rare ion channel mutations associated with familial atrial fibrillation. J. Am. Coll. Cardiol. 60, 1173–1181 (2012).
Otway, R. et al. Stretch-sensitive KCNQ1 mutation: a link between genetic and environmental factors in the pathogenesis of atrial fibrillation? J. Am. Coll. Cardiol. 49, 578–586 (2007).
Gudbjartsson, D. F. et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 488, 353–357 (2007).
Kaab, S. et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur. Heart J. 30, 813–819 (2009).
Body, S. C. et al. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ. Cardiovasc. Genet. 2, 499–506 (2009).
Gudbjartsson, D. F. et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat. Genet. 8, 876–878 (2009).
Benjamin, E. J. et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat. Genet. 41, 879–881 (2009).
Ellinor, P. T. et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat. Genet. 42, 240–244 (2010).
Ellinor, P. T. et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat. Genet. 44, 670–675 (2012).
Nattel, S. New ideas about atrial fibrillation 50 years on. Nature 415, 219–226 (2002).
Xia, M. et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem. Biophys. Res. Commun. 332, 1012–1019 (2005).
Yang, Y. et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am. J. Hum. Genet. 75, 899–905 (2004).
Yang, T., Yang, P., Roden, D. M. & Darbar, D. Novel KCNA5 mutation implicates tyrosine kinase signaling in human atrial fibrillation. Heart Rhythm 7, 1246–1252 (2010).
Shinlapawittayatorn, K. & Deschenes, I. Alteration of tyrosine kinase signaling: another player in the arrhythmogenesis of atrial fibrillation? Heart Rhythm 7, 1253–1254 (2010).
Chung, M. K. et al. C-reactive protein elevation in patients with atrial arrhythmias: inflammatory mechanisms and persistence of atrial fibrillation. Circulation 104, 2886–2891 (2001).
Vollmar, A. M. The role of atrial natriuretic peptide in the immune system. Peptides 26, 1086–1094 (2005).
Roberts, J. D. & Gollob, M. H. Impact of genetic discoveries on the classification of lone atrial fibrillation. J. Am. Coll. Cardiol. 55, 705–712 (2010).
Li, D., Fareh, S., Leung, T. K. & Nattel, S. Promotion of atrial fibrillation by heart failure in dogs: atrial remodeling of a different sort. Circulation 100, 87–95 (1999).
Disertori, M. et al. Autosomal recessive atrial dilated cardiomyopathy with standstill evolution associated with mutation of natriuretic peptide precursor A. Circ. Cardiovasc. Genet. 6, 27–36 (2013).
Savio-Galimberti, E. et al. NPPA overexpression in mice increases susceptibility to atrial fibrillation [abstract]. Circulation 126, A19074 (2012).
Olson, T. M. et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum. Mol. Genet. 15, 2185–2191 (2006).
Yang, Y. et al. Novel KCNA5 loss-of-function mutations responsible for atrial fibrillation. J. Hum. Genet. 54, 277–283 (2009).
Christophersen, I. E. et al. Genetic variation in KCNA5: impact on the atrial-specific potassium current IKur in patients with lone atrial fibrillation. Eur. Heart J. http://dx.doi.org/10.1093/eurheartj/ehs442.
Satoh, T. & Zipes, D. P. Cesium-induced atrial tachycardia degenerating into atrial fibrillation in dogs: atrial torsades de pointes? J. Cardiovasc. Electrophysiol. 9, 970–975 (1998).
Ehrlich, J. R., Zicha, S., Coutu, P., Hebert, T. E. & Nattel, S. Atrial fibrillation-associated minK38G/S. polymorphism modulates delayed rectifier current and membrane localization. Cardiovasc. Res. 67, 520–528 (2005).
Johnson, J. N. et al. Prevalence of early-onset atrial fibrillation in congenital long QT syndrome. Heart Rhythm 5, 704–709 (2008).
Lemoine, M. D. et al. Arrhythmogenic left atrial cellular electrophysiology in a murine genetic long QT syndrome model. Cardiovasc. Res. 92, 67–74 (2011).
Gollob, M. H. et al. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N. Engl. J. Med. 354, 2677–2688 (2006).
Firouzi, M. et al. Association of human connexin 40 gene polymorphisms with atrial vulnerability as a risk factor for idiopathic atrial fibrillation. Circ. Res. 95, e29–e33 (2004).
Firouzi, M. et al. The human Cx40 promoter polymorphism −44G A differentially affects transcriptional regulation by Sp1 and GATA4. Biochim. Biophys. Acta 1759, 491–496 (2006).
Yang, Y. Q. et al. Novel connexin40 missense mutations in patients with familial atrial fibrillation. Europace 12, 1421–1427 (2010).
Yang, Y. Q. et al. Connexin40 nonsense mutation in familial atrial fibrillation. Int. J. Mol. Med. 26, 605–610 (2010).
Gu, J. Y., Xu, J. H., Yu, H. & Yang, Y. Q. Novel GATA5 loss-of-function mutations underlie familial atrial fibrillation. Clinics (Sao Paulo) 67, 1393–1399 (2012).
Sun, Y. et al. Novel germline GJA5/connexin40 mutations associated with lone atrial fibrillation impair gap junctional intercellular communication. Hum. Mutat. 34, 603–609 (2013).
Christophersen, I. E. et al. Rare variants in GJA5 are associated with early-onset lone atrial fibrillation. Can. J. Cardiol. 29, 111–116 (2013).
Muller, I. I. et al. Use of a zebrafish model demonstrates that atrial fibrillation-associated gene GREM2 regulates cardiac laterality, cardiomyocyte differentiation and atrial rhythm. Dis. Model. Mech. 6, 332–341 (2013).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Lubitz, S. A. et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation 122, 976–984.
Mommersteeg, M. T. et al. Pitx2c and Nkx2–5 are required for the formation and identity of the pulmonary myocardium. Circ. Res. 101, 902–909 (2007).
Wang, J. et al. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl Acad. Sci. USA 107, 9753–9758 (2010).
Kirchhof, P. et al. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ. Cardiovasc. Genet. 4, 123–133 (2011).
Chinchilla, A. et al. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ. Cardiovasc. Genet. 4, 269–279 (2011).
Ihida-Stansbury, K. et al. Paired-related homeobox gene Prx1 is required for pulmonary vascular development. Circ. Res. 94, 1507–1514 (2004).
Haissaguerre, M. et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N. Engl. J. Med. 339, 659–666 (1998).
Pandit, S. V. et al. Ionic determinants of functional reentry in a 2-D model of human atrial cells during simulated chronic atrial fibrillation. Biophys. J. 88, 3806–3821 (2005).
Voigt, N. et al. Left-to-right atrial inward rectifier potassium current gradients in patients with paroxysmal versus chronic atrial fibrillation. Circ. Arrhythm. Electrophysiol. 3, 472–480 (2010).
Volonte, D., McTiernan, C. F., Drab, M., Kasper, M. & Galbiati, F. Caveolin-1 and caveolin-3 form heterooligomeric complexes in atrial cardiac myocytes that are required for doxorubicin-induced apoptosis. Am. J. Physiol. Heart Circ. Physiol. 294, H392–H401 (2008).
Stieber, J. et al. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc. Natl Acad. Sci. USA 100, 15235–15240 (2003).
Nof, E. et al. Point mutation in the HCN4 cardiac ion channel pore affecting synthesis, trafficking, and functional expression is associated with familial asymptomatic sinus bradycardia. Circulation 116, 463–470 (2007).
Zhao, Y. Y. et al. Defects in caveolin-1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proc. Natl Acad. Sci. USA 99, 11375–11380 (2002).
Sinner, M. F. et al. The non-synonymous coding IKr-channel variant KCNH2-K897T is associated with atrial fibrillation: results from a systematic candidate gene-based analysis of KCNH2 (HERG). Eur. Heart J. 29, 907–914 (2008).
Berry, F. B. et al. Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J. Biol. Chem. 276, 25057–25065 (2001).
Sun, X. et al. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat. Genet. 37, 407–412 (2005).
Burgner, D. et al. A genome-wide association study identifies novel and functionally related susceptibility loci for Kawasaki disease. PLoS Genet. 5, e1000319 (2009).
Lubitz, S. A. et al. Genetics of atrial fibrillation: implications for future research directions and personalized medicine. Circ. Arrhythm. Electrophysiol. 3, 291–299 (2010).
Qi, Y. et al. Atbf1 is required for the Pit1 gene early activation. Proc. Natl Acad. Sci. USA 105, 2481–2486 (2008).
Kim, T. S. et al. The ZFHX3 (ATBF1) transcription factor induces PDGFRB, which activates ATM in the cytoplasm to protect cerebellar neurons from oxidative stress. Dis. Model. Mech. 3, 752–762 (2010).
Verheule, S. et al. Increased vulnerability to atrial fibrillation in transgenic mice with selective atrial fibrosis caused by overexpression of TGF-β1. Circ. Res. 94, 1458–1465 (2004).
Mabuchi, M. et al. Tumor suppressor, AT motif binding factor 1 (ATBF1), translocates to the nucleus with runt domain transcription factor 3 (RUNX3) in response to TGF-β signal transduction. Biochem. Biophys. Res. Commun. 398, 321–325 (2010).
Li, J. et al. Role of inflammation and oxidative stress in atrial fibrillation. Heart Rhythm 7, 438–444 (2010).
Austin, E. D. et al. Whole exome sequencing to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ. Cardiovasc. Genet. 5, 336–343 (2012).
Del Galdo, F., Lisanti, M. P. & Jimenez, S. A. Caveolin-1, transforming growth factor-beta receptor internalization, and the pathogenesis of systemic sclerosis. Curr. Opin. Rheumatol. 20, 713–719 (2008).
Cohen, R. I. et al. Ghrelin receptor expression in lymphocytes isolated from adult cystic fibrosis patients. Respiration 79, 141–146 (2009).
Fritz, D. & Stefanovic, B. RNA-binding protein RBMS3 is expressed in activated hepatic stellate cells and liver fibrosis and increases expression of transcription factor Prx1. J. Mol. Biol. 371, 585–595 (2007).
reSOLVE. Wound Healing and Fibrosis-related Genes [online], (2013).
Beqqali, A. et al. CHAP is a newly identified Z-disc protein essential for heart and skeletal muscle function. J. Cell Sci. 123, 1141–1150 (2010).
Homer, R. J. & Herzog, E. L. Recent advances in pulmonary fibrosis: implications for scleroderma. Curr. Opin. Rheumatol. 22, 683–689 (2010).
He, W. et al. Wnt/β-catenin signaling promotes renal interstitial fibrosis. J. Am. Soc. Nephrol. 20, 765–776 (2009).
Ellinor, P. T. et al. Mutations in the long QT gene, KCNQ1, are an uncommon cause of atrialfibrillation. Heart 90, 1487–1488 (2004).
Darbar, D., Motsinger, A. A., Ritchie, M. D., Gainer, J. V. & Roden, D. M. Polymorphism modulates symptomatic response to antiarrhythmic drug therapy in patients with lone atrial fibrillation. Heart Rhythm 4, 743–749 (2007).
Roden, D. M. et al. Pharmacogenomics: challenges and opportunities. Ann. Intern. Med. 145, 749–757 (2006).
Giacomini, K. M. et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin. Pharmacol. Ther. 81, 328–345 (2007).
US National Library of Medicine. ClinicalTrials.gov [online], (2013).
Savelieva, I. & Camm, A. J. Clinical relevance of silent atrial fibrillation: prevalence, prognosis, quality of life, and management. J. Interv. Card. Electrophysiol. 4, 369–382 (2000).
Friedman, P. A. et al. Atrial therapies reduce atrial arrhythmia burden in defibrillator patients. Circulation 104, 1023–1028 (2001).
Israel, C. W., Grönefeld, G., Ehrlich, J. R., Li, Y. G. & Hohnloser, S. H. Long-term risk of recurrent atrial fibrillation as documented by an implantable monitoring device: implications for optimal patient care. J. Am. Coll. Cardiol. 43, 47–52 (2004).
Euler, D. E. & Friedman, P. A. Atrial arrhythmia burden as an endpoint in clinical trials: is it the best surrogate? Lessons from a multicenter defibrillator trial. Card. Electrophysiol. Rev. 7, 355–358 (2003).
Charitos, E. I. et al. A comprehensive evaluation of rhythm monitoring strategies for the detection of atrial fibrillation recurrence: insights from 647 continuously monitored patients and implications for monitoring after therapeutic interventions. Circulation 126, 806–814 (2012).
Hindricks, G. & Piorkowski, C. Atrial fibrillation monitoring: mathematics meets real life. Circulation 126, 791–792 (2012).
Calkins, H. et al. 2012 HRS/EHRA/ECAS expert consensus statement on catheter and surgical ablation of atrial fibrillation: recommendations for patient selection, procedural techniques, patient management and follow-up, definitions, endpoints, and research trial design: a report of the Heart Rhythm Society (HRS) Task Force on Catheter and Surgical Ablation of Atrial Fibrillation. Developed in partnership with the European Heart Rhythm Association (EHRA), a registered branch of the European Society of Cardiology (ESC) and the European Cardiac Arrhythmia Society (ECAS); and in collaboration with the American College of Cardiology (ACC), American Heart Association (AHA), the Asia Pacific Heart Rhythm Society (APHRS), and the Society of Thoracic Surgeons (STS). Endorsed by the governing bodies of the American College of Cardiology Foundation, the American Heart Association, the European Cardiac Arrhythmia Society, the European Heart Rhythm Association, the Society of Thoracic Surgeons, the Asia Pacific Heart Rhythm Society, and the Heart Rhythm Society. Heart Rhythm 9, 632–696.e621 (2012).
Parvez, B. et al. Symptomatic response to antiarrhythmic drug therapy is modulated by a common single nucleotide polymorphism in atrial fibrillation. J. Am. Coll. Cardiol. 60, 539–545 (2012).
Nia, A. M. et al. β1-Adrenoceptor polymorphism predicts flecainide action in patients with atrial fibrillation. PLoS ONE 5, e11421 (2010).
Parvez, B. et al. A common β1-adrenergic receptor polymorphism predicts favorable response to rate-control therapy in atrial fibrillation. J. Am. Coll. Cardiol. 59, 49–56 (2012).
Husser, D., Adams, V., Piorkowski, C., Hindricks, G. & Bollmann, A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J. Am. Coll. Cardiol. 55, 747–753 (2010).
Shoemaker, B. M. et al. Common atrial fibrillation risk alleles at 4q25 predict recurrence after catheter-based atrial fibrillation ablation. Heart Rhythm 10, 394–400 (2013).
Parvez, B. et al. Common genetic polymorphisms at 4q25 locus predicts predicts atrial fibrillation after successful cardioversion. Heart Rhythm http://dx.doi.org/10.1016/j.hrthm.2013.02.018.
Olshansky, B. et al. The atrial fibrillation follow-up investigation of rhythm management (AFFIRM) study: approaches to control rate in atrial fibrillation. J. Am. Coll. Cardiol. 43, 1201–1208 (2004).
Muhammad, R. et al. Genome-wide association analysis to identify genomic modulators of rate control therapy in patients with atrial fibrillation [abstract]. Circulation 124, A12933 (2011).
Pfeufer, A. et al. Genome-wide association study of PR interval. Nat. Genet. 42, 153–159 (2010).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Parvez, B. & Darbar, D. The “missing” link in atrial fibrillation heritability. J. Electrocardiol. 44, 641–644 (2011).
Abraham, R. L., Yang, T., Blair, M., Roden, D. M. & Darbar, D. Augmented potassium current is a shared phenotype for two genetic defects associated with familial atrial fibrillation. J. Mol. Cell. Cardiol. 48, 181–190 (2010).
Hong, K. et al. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc. Res. 68, 433–440 (2005).
Lundby, A., Ravn, L. S., Svendsen, J. H., Olesen, S. P. & Schmitt, N. KCNQ1 mutation Q147R is associated with atrial fibrillation and prolonged QT interval. Heart Rhythm 4, 1532–1541 (2007).
Bartos, D. C. et al. A KCNQ1 mutation causes a high penetrance for familial atrial fibrillation. J. Cardiovasc. Electrophysiol. http://dx.doi.org/10.1111/jce.12068.
Henrion, U. et al. Overlapping cardiac phenotype associated with a familial mutation in the voltage sensor of the KCNQ1 channel. Cell. Physiol. Biochem. 29, 809–818 (2012).
Bartos, D. C. et al. R231C mutation in KCNQ1 causes long QT syndrome type 1 and familial atrial fibrillation. Heart Rhythm 8, 48–55 (2011).
Das, S. et al. Mutation in the S3 segment of KCNQ1 results in familial lone atrial fibrillation. Heart Rhythm 6, 1146–1153 (2009).
Olesen, M. S. et al. Mutations in the potassium channel subunit KCNE1 are associated with early-onset familial atrial fibrillation. BMC Med. Genet. 13, 24 (2012).
Ravn, L. S. et al. Gain of function in IKs secondary to a mutation in KCNE5 associated with atrial fibrillation. Heart Rhythm 5, 427–435 (2008).
Kharche, S. et al. Atrial proarrhythmia due to increased inward rectifier current (IK1) arising from KCNJ2 mutation—a simulation study. Prog. Biophys. Mol. Biol. 98, 186–197 (2008).
Hattori, T. et al. A novel gain-of-function KCNJ2 mutation associated with short-QT syndrome impairs inward rectification of Kir2.1 currents. Cardiovasc. Res. 93, 666–673 (2012).
Shi, H. F. et al. Prevalence and spectrum of GJA5 mutations associated with lone atrial fibrillation. Mol. Med. Rep. 7, 767–774 (2013).
Laitinen-Forsblom, P. J. et al. SCN5A mutation associated with cardiac conduction defect and atrial arrhythmias. J. Cardiovasc. Electrophysiol. 17, 480–485 (2006).
Ellinor, P. T. et al. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm 5, 99–105 (2008).
Darbar, D. et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation 117, 1927–1935 (2008).
Makiyama, T. et al. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J. Am. Coll. Cardiol. 52, 1326–1334 (2008).
Li, Q. et al. Gain-of-function mutation of Nav1.5 in atrial fibrillation enhances cellular excitability and lowers the threshold for action potential firing. Biochem. Biophys. Res. Commun. 380, 132–137 (2009).
Watanabe, H. et al. Mutations in sodium channel β1- and β2-subunits associated with atrial fibrillation. Circ. Arrhythm. Electrophysiol. 2, 268–275 (2009).
Wang, P. et al. Functional dominant-negative mutation of sodium channel subunit gene SCN3B associated with atrial fibrillation in a Chinese GeneID population. Biochem. Biophys. Res. Commun. 398, 98–104 (2010).
Olesen, M. S. et al. Mutations in sodium channel β-subunit SCN3B are associated with early-onset lone atrial fibrillation. Cardiovasc. Res. 89, 786–793 (2010).
Olesen, M. S., Holst, A. G., Svendsen, J. H., Haunso, S. & Tfelt-Hansen, J. SCN1Bb R214Q found in 3 patients: 1 with Brugada syndrome and 2 with lone atrial fibrillation. Heart Rhythm 9, 770–773 (2012).
Olesen, M. S. et al. High prevalence of long QT syndrome-associated SCN5A variants in patients with early-onset lone atrial fibrillation. Circ. Cardiovasc. Genet. 5, 450–459 (2012).
Muhammad, R. et al. Rare SCN10A variants modulate ventricular response rates during atrial fibrillation [abstract]. Circulation 124, A13007 (2011).
Splawski, I. et al. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc. Natl Acad. Sci. USA 102, 8089–8968 (2005).
Antzelevitch, C. et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation 115, 442–449 (2007).
Ren, X. et al. Identification of NPPA variants associated with atrial fibrillation in a Chinese GeneID population. Clin. Chim. Acta 411, 481–485 (2010).
Roberts, J. D. et al. Evaluation of non-synonymous NPPA single nucleotide polymorphisms in atrial fibrillation. Europace 12, 1078–1083 (2010).
Zhang, X. et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 135, 1017–1027 (2008).
Acknowledgements
This work was supported by NIH grants (U19 HL65962 and R01 HL092217), and an AHA Established Investigator Award (0940116N).
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article and contributed substantially to discussions of its content. D. Darbar wrote the manuscript, and both authors reviewed/edited it before submission.
Corresponding author
Ethics declarations
Competing interests
Dawood Darbar has received grant or research support from the AHA and the NIH. Dan M. Roden receives royalties from Clinical Data, Inc. on a patent to predict drug-induced arrhythmias.
Rights and permissions
About this article
Cite this article
Darbar, D., Roden, D. Genetic mechanisms of atrial fibrillation: impact on response to treatment. Nat Rev Cardiol 10, 317–329 (2013). https://doi.org/10.1038/nrcardio.2013.53
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrcardio.2013.53
This article is cited by
-
Transcriptional factors in calcium mishandling and atrial fibrillation development
Pflügers Archiv - European Journal of Physiology (2021)
-
Big Data and Atrial Fibrillation: Current Understanding and New Opportunities
Journal of Cardiovascular Translational Research (2020)
-
Genome-wide association study of myocardial infarction, atrial fibrillation, acute stroke, acute kidney injury and delirium after cardiac surgery – a sub-analysis of the RIPHeart-Study
BMC Cardiovascular Disorders (2019)
-
Ionic and cellular mechanisms underlying TBX5/PITX2 insufficiency-induced atrial fibrillation: Insights from mathematical models of human atrial cells
Scientific Reports (2018)
-
Gene-guided therapy for catheter-ablation of atrial fibrillation: are we there yet?
Journal of Interventional Cardiac Electrophysiology (2016)