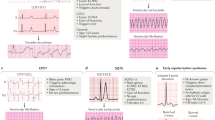
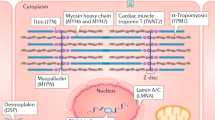
Abstract
Over the past 2 decades, investigators in the field of cardiac genetics have evolved a complex understanding of the pathophysiological basis of inherited cardiac diseases, which predispose individuals to sudden cardiac death. In this Review, we describe the current status of gene discovery and the associations between phenotype and genotype in the cardiac channelopathies and cardiomyopathies. The various indications for genetic testing and its utility in the clinic are assessed in relation to diagnosis, cascade testing, guiding management, and prognosis. Some common problems exist across all phenotypes: the variable penetrance and expressivity of genetic disease, and the difficulty of assessing the functional and clinical effects of novel mutations. These issues will be of particular importance as the next-generation sequencing technologies are used by genetics laboratories to provide results from large panels of genes. The accurate interpretation of these results will be the main challenge for the future.
Key Points
-
Genetic testing for cardiac channelopathies and cardiomyopathies has developed substantially over the past 2 decades and is a potentially useful tool for clinicians, if used appropriately
-
Variable penetrance and expressivity of genetic disease are common and, combined with 'variants of unknown significance', complicate the interpretation of the results of genetic testing
-
The yield of genetic testing is never 100% for any given phenotype; as a diagnostic tool, therefore, genetic testing is largely limited to confirmation of disease
-
Genetic testing is, however, particularly useful in families in which a 'true' disease-causing mutation is found, and can be used to identify carriers and to reassure noncarriers
-
Genetic testing can be especially helpful in guiding therapy and assessing prognosis for long QT syndrome, but not in many other conditions
-
The future holds promise, but also challenges, for the interpretation of variants of unknown significance and the huge amount of genetic data that will be produced by next-generation sequencing technologies
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Geisterfer-Lowrance, A. A. et al. A molecular basis for familial hypertrophic cardiomyopathy: a β cardiac myosin heavy chain gene missense mutation. Cell 62, 999–1006 (1990).
Wang, Q. et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell 80, 805–811 (1995).
Curran, M. E. et al. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 80, 795–803 (1995).
van der Werf, C. et al. Diagnostic yield in sudden unexplained death and aborted cardiac arrest in the young: the experience of a tertiary referral center in the Netherlands. Heart Rhythm 7, 1383–1389 (2010).
Behr, E. R. et al. Sudden arrhythmic death syndrome: familial evaluation identifies inheritable heart disease in the majority of families. Eur. Heart J. 29, 1670–1680 (2008).
Hofman, N. et al. Active cascade screening in primary inherited arrhythmia syndromes, does it lead to prophylactic treatment? J. Am. Coll. Cardiol. 55, 2570–2576 (2010).
Schwartz, P. J., Moss, A. J., Vincent, G. M. & Crampton, R. S. Diagnostic criteria for the long QT syndrome: an update. Circulation 88, 782–784 (1993).
Hedley, P. L. et al. The genetic basis of long QT and short QT syndromes: a mutation update. Hum. Mutat. 30, 1486–1511 (2009).
Priori, S. G., Napolitano, C. & Schwartz, P. J. Low penetrance in the long-QT syndrome: clinical impact. Circulation 99, 529–533 (1999).
Ackerman, M. J. et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace 13, 1077–1109 (2011).
Schwartz, P. J. et al. Genotype–phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation 103, 89–95 (2001).
Zhang, L. et al. Spectrum of ST-T-wave patterns and repolarization parameters in congenital long-QT syndrome: ECG findings identify genotypes. Circulation 102, 2849–2855 (2000).
Sauer, A. J. et al. Long QT syndrome in adults. J. Am. Coll. Cardiol. 49, 329–337 (2007).
Goldenberg, I. et al. Risk for life-threatening cardiac events in patients with genotype-confirmed long-QT syndrome and normal-range corrected QT intervals. J. Am. Coll. Cardiol. 57, 51–59 (2011).
Priori, S. G. et al. Risk stratification in the long-QT syndrome. N. Engl. J. Med. 348, 1866–1874 (2003).
Goldenberg, I. et al. Risk factors for aborted cardiac arrest and sudden cardiac death in children with the congenital long-QT syndrome. Circulation 117, 2184–2191 (2008).
Liu, J. F. et al. Risk factors for recurrent syncope and subsequent fatal or near-fatal events in children and adolescents with long QT syndrome. J. Am. Coll. Cardiol. 57, 941–950 (2011).
Seth, R. et al. Long QT syndrome and pregnancy. J. Am. Coll. Cardiol. 49, 1092–1098 (2007).
Moss, A. J. et al. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation 115, 2481–2489 (2007).
Moss, A. J. et al. Increased risk of arrhythmic events in long-QT syndrome with mutations in the pore region of the human ether-a-go-go-related gene potassium channel. Circulation 105, 794–799 (2002).
Westenskow, P., Splawski, I., Timothy, K. W., Keating, M. T. & Sanguinetti, M. C. Compound mutations: a common cause of severe long-QT syndrome. Circulation 109, 1834–1841 (2004).
Tomas, M. et al. Polymorphisms in the NOS1AP gene modulate QT interval duration and risk of arrhythmias in the long QT syndrome. J. Am. Coll. Cardiol. 55, 2745–2752 (2010).
Crotti, L. et al. NOS1AP is a genetic modifier of the long-QT syndrome. Circulation 120, 1657–1663 (2009).
Amin, A. S. et al. Variants in the 3' untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. Eur. Heart J. 33, 714–723 (2012).
Crotti, L. et al. KCNH2-K897T is a genetic modifier of latent congenital long-QT syndrome. Circulation 112, 1251–1258 (2005).
Roden, D. M. & Yang, T. Protecting the heart against arrhythmias: potassium current physiology and repolarization reserve. Circulation 112, 1376–1378 (2005).
CredibleMeds™. Drugs lists by risk groups: Drugs that prolong the QT interval and/or induce torsades de pointes [online], (2013).
Itoh, H. et al. Latent genetic backgrounds and molecular pathogenesis in drug-induced long-QT syndrome. Circ. Arrhythm. Electrophysiol. 2, 511–523 (2009).
Paulussen, A. D. et al. Genetic variations of KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2 in drug-induced long QT syndrome patients. J. Mol. Med. (Berl.) 82, 182–188 (2004).
Yang, P. et al. Allelic variants in long-QT disease genes in patients with drug-associated torsades de pointes. Circulation 105, 1943–1948 (2002).
Splawski, I. et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science 297, 1333–1336 (2002).
Kaab, S. et al. A large candidate gene survey identifies the KCNE1 D85N polymorphism as a possible modulator of drug-induced torsades de pointes. Circ. Cardiovasc. Genet. 5, 91–99 (2012).
Jamshidi, Y. et al. Common variation in the NOS1AP gene is associated with drug-induced QT prolongation and ventricular arrhythmia. J. Am. Coll. Cardiol. 60, 841–850 (2012).
Goldenberg, I. et al. β-Blocker efficacy in high-risk patients with the congenital long-QT syndrome types 1 and 2: implications for patient management. J. Cardiovasc. Electrophysiol. 21, 893–901 (2010).
Barsheshet, A. et al. Mutations in cytoplasmic loops of the KCNQ1 channel and the risk of life-threatening events: implications for mutation-specific response to beta blocker therapy in type 1 long-QT syndrome. Circulation 125, 1988–1996 (2012).
Etheridge, S. P., Compton, S. J., Tristani-Firouzi, M. & Mason, J. W. A new oral therapy for long QT syndrome: long-term oral potassium improves repolarization in patients with HERG mutations. J. Am. Coll. Cardiol. 42, 1777–1782 (2003).
Makita, N. et al. The E1784K mutation in SCN5A is associated with mixed clinical phenotype of type 3 long QT syndrome. J. Clin. Invest. 118, 2219–2229 (2008).
Remme, C. A. & Wilde, A. A. Late sodium current inhibition in acquired and inherited ventricular (dys)function and arrhythmias. Cardiovasc. Drugs Ther. 27, 91–101 (2013).
Behr, E. R. & Roden, D. M. Drug-induced arrhythmia: pharmacogenomic prescribing? Eur. Heart J. 34, 89–95 (2013).
Antzelevitch, C. et al. Brugada syndrome: report of the second consensus conference: endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation 111, 659–670 (2005).
Govindan, M. et al. Utility of high and standard right precordial leads during ajmaline testing for the diagnosis of Brugada syndrome. Heart 96, 1904–1908 (2010).
Schulze-Bahr, E. et al. Sodium channel gene (SCN5A) mutations in 44 index patients with Brugada syndrome: different incidences in familial and sporadic disease. Hum. Mutat. 21, 651–652 (2003).
Mizusawa, Y. & Wilde, A. A. M. Arrhythmogenic disorders of genetic origin: Brugada syndrome. Circ. Arrhythm. Electrophysiol. 5, 606–616 (2012).
Bastiaenen, R. & Behr, E. R. Sudden death and ion channel disease: pathophysiology and implications for management. Heart 97, 1365–1372 (2011).
Kapplinger, J. D. et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm 7, 33–46 (2010).
Bezzina, C. et al. A single Na+ channel mutation causing both long-QT and Brugada syndromes. Circ. Res. 85, 1206–1213 (1999).
Antzelevitch, C. et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation 115, 442–449 (2007).
Crotti, L. et al. Spectrum and prevalence of mutations involving BrS1- through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J. Am. Coll. Cardiol. 60, 1410–1418 (2012).
Probst, V. et al. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome. Circ. Cardiovasc. Genet. 2, 552–557 (2009).
Meregalli, P. G. et al. Type of SCN5A mutation determines clinical severity and degree of conduction slowing in loss-of-function sodium channelopathies. Heart Rhythm 6, 341–348 (2009).
Probst, V. et al. Long-term prognosis of patients diagnosed with Brugada syndrome: results from the FINGER Brugada syndrome registry. Circulation 121, 635–643 (2010).
Nishii, N., Ogawa, M. & Morita, H. et al. SCN5A mutation is associated with early and frequent recurrence of ventricular fibrillation in patients with Brugada syndrome. Circ. J. 74, 2572–2578 (2010).
Bezzina, C. R. et al. Common sodium channel promoter haplotype in Asian subjects underlies variability in cardiac conduction. Circulation 113, 338–344 (2006).
Leenhardt, A. et al. Catecholaminergic polymorphic ventricular tachycardia in children: a 7-year follow-up of 21 patients. Circulation 91, 1512–1519 (1995).
Priori, S. G. et al. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103, 196–200 (2001).
Laitinen, P. J. et al. Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation 103, 485–490 (2001).
Medeiros-Domingo, A. et al. Comprehensive open reading frame mutational analysis of the RYR2-encoded ryanodine receptor/calcium channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome. J. Am. Coll. Cardiol. 54, 2065–2074 (2009).
Lahat, H. et al. A missense mutation in a highly conserved region of CASQ2 is associated with autosomal recessive catecholamine-induced polymorphic ventricular tachycardia in Bedouin families from Israel. Am. J. Hum. Genet. 69, 1378–1384 (2001).
Cerrone, M., Napolitano, C. & Priori, S. G. Catecholaminergic polymorphic ventricular tachycardia: a paradigm to understand mechanisms of arrhythmias associated to impaired Ca2+ regulation. Heart Rhythm 6, 1652–1659 (2009).
Nyegaard, M. et al. Mutations in calmodulin cause ventricular tachycardia and sudden cardiac death. Am. J. Hum. Genet. 91, 703–712 (2012).
Roux-Buisson, N. et al. Absence of triadin, a protein of the calcium release complex, is responsible for cardiac arrhythmia with sudden death in human. Hum. Mol. Genet. 21, 2759–2767 (2012).
Tester, D. J. et al. Genotypic heterogeneity and phenotypic mimicry among unrelated patients referred for catecholaminergic polymorphic ventricular tachycardia genetic testing. Heart Rhythm 3, 800–805 (2006).
Priori, S. G. et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation 106, 69–74 (2002).
Hayashi, M. et al. Incidence and risk factors of arrhythmic events in catecholaminergic polymorphic ventricular tachycardia. Circulation 119, 2426–2434 (2009).
van der Werf, C. et al. Familial evaluation in catecholaminergic polymorphic ventricular tachycardia: disease penetrance and expression in cardiac ryanodine receptor mutation-carrying relatives. Circ. Arrhythm. Electrophysiol. 5, 748–756 (2012).
Brugada, R. et al. Sudden death associated with short-QT syndrome linked to mutations in HERG. Circulation 109, 30–35 (2004).
Bellocq, C. et al. Mutation in the KCNQ1 gene leading to the short QT-interval syndrome. Circulation 109, 2394–2397 (2004).
Priori, S. G. et al. A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ. Res. 96, 800–807 (2005).
Haïssaguerre, M. et al. Ventricular fibrillation with prominent early repolarization changes associated to a rare variant of KCNJ8/KATP channel. J. Cardiovasc. Electrophysiol. 20, 93–98 (2009).
Watanabe, H. et al. Electrocardiographic characteristics and SCN5A mutations in idiopathic ventricular fibrillation associated with early repolarization. Circ. Arrhythm. Electrophysiol. 4, 874–881 (2011).
Burashnikov, E. et al. Mutations in the cardiac L-type calcium channel associated with inherited J-wave syndromes and sudden cardiac death. Heart Rhythm 7, 1872–1882 (2010).
Kruse, M. et al. Impaired endocytosis of the ion channel TRPM4 is associated with human progressive familial heart block type I. J. Clin. Invest. 119, 2737–2744 (2009).
Schott, J. J. et al. Cardiac conduction defects associate with mutations in SCN5A. Nat. Genet. 23, 20–21 (1999).
Alders, M. et al. Haplotype sharing analysis implicates chromosome 7q36 harboring DPP6 in familial idiopathic ventricular fibrillation. Am. J. Hum. Genet. 84, 468–476 (2009).
Chen, Y. H. et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science 299, 251–254 (2003).
Yang, Y. et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am. J. Hum. Genet. 75, 899–905 (2004).
Xia, M. et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem. Biophys. Res. Commun. 332, 1012–1019 (2005).
Bartos, D. C. et al. R231C mutation in KCNQ1 causes long QT syndrome type 1 and familial atrial fibrillation. Heart Rhythm 8, 48–55 (2011).
Schulze-Bahr, E. et al. Pacemaker channel dysfunction in a patient with sinus node disease. J. Clin. Invest. 111, 1537–1545 (2003).
Benson, D. W. et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A). J. Clin. Invest. 112, 1019–1028 (2003).
Laurent, G. et al. Multifocal ectopic Purkinje-related premature contractions: a new SCN5A-related cardiac channelopathy. J. Am. Coll. Cardiol. 60, 144–156 (2012).
Saffitz, J. E. The pathobiology of arrhythmogenic cardiomyopathy. Annu. Rev. Pathol. 6, 299–321 (2011).
Gersch, B. J. et al. ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 58, e212–e260 (2011).
Christiaans, I. et al. Manifest disease, risk factors for sudden cardiac death, and cardiac events in a large nationwide cohort of predictively tested hypertrophic cardiomyopathy mutation carriers: determining the best cardiological screening strategy. Eur. Heart J. 32, 1161–1170 (2011).
Christiaans, I. et al. Risk stratification for sudden cardiac death in hypertrophic cardiomyopathy: systematic review of clinical risk markers. Europace 12, 313–321 (2010).
Watkins, H., Thierfelder, L. & Hwang, D. S. et al. Sporadic hypertrophic cardiomyopathy due to de novo myosin mutations. J. Clin. Invest. 90, 1666–1671 (1992).
Watkins, H. et al. Mutations in the genes for cardiac troponin T and α-tropomyosin in hypertrophic cardiomyopathy. N. Engl. J. Med. 332, 1058–1064 (1995).
Watkins, H. et al. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N. Engl. J. Med. 326, 1108–1114 (1992).
Varnava, A. M. et al. Hypertrophic cardiomyopathy: histopathological features of sudden death in cardiac troponin T disease. Circulation 104, 1380–1384 (2001).
Pasquale, F. et al. Long-term outcomes in hypertrophic cardiomyopathy caused by mutations in the cardiac troponin T gene. Circ. Cardiovasc. Genet. 5, 10–17 (2012).
Richard, P. et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 107, 2227–2232 (2003).
Van Driest, S. L., Ommen, S. R., Tajik, A. J., Gersh, B. J. & Ackerman, M. J. Yield of genetic testing in hypertrophic cardiomyopathy. Mayo Clin. Proc. 80, 739–744 (2005).
Mestroni, L. et al. Guidelines for the study of familial dilated cardiomyopathies: collaborative research group of the European Human and Capital Mobility Project on familial dilated cardiomyopathy. Eur. Heart J. 20, 93–102 (1999).
van Spaendonck-Zwarts, K. Y. et al. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years' experience. Eur. J. Heart Fail. 15, 628–636 (2013).
Herman, D. S. et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 366, 619–628 (2012).
van Rijsingen, I. A. et al. Risk factors for malignant ventricular arrhythmias in lamin A/C mutation carriers: a European cohort study. J. Am. Coll. Cardiol. 59, 493–500 (2012).
Basso, C., Corrado, D., Bauce, B. & Thiene, G. Arrhythmogenic right ventricular cardiomyopathy. Circ. Arrhythm. Electrophysiol. 5, 1233–1246 (2012).
Kapplinger, J. D. et al. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J. Am. Coll. Cardiol. 57, 2317–2327 (2011).
Al-Jassar, C. et al. The nonlinear structure of the desmoplakin plakin domain and the effects of cardiomyopathy-linked mutations. J. Mol. Biol. 411, 1049–1061 (2011).
Quarta, G. et al. Familial evaluation in arrhythmogenic right ventricular cardiomyopathy: impact of genetics and revised task force criteria. Circulation 123, 2701–2709 (2011).
Cox, M. G. et al. Arrhythmogenic right ventricular dysplasia/cardiomyopathy: pathogenic desmosome mutations in index-patients predict outcome of family screening: Dutch Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Genotype–Phenotype Follow-up Study. Circulation 123, 2690–2700 (2011).
Semsarian, C. & Hamilton, R. M. Key role of the molecular autopsy in sudden unexpected death. Heart Rhythm 9, 145–150 (2012).
Raju, H. & Behr, E. R. Unexplained sudden death, focussing on genetics and family phenotyping. Curr. Opin. Cardiol. 28, 19–25 (2013).
Giudicessi, J. R. et al. Phylogenetic and physicochemical analyses enhance the classification of rare nonsynonymous single nucleotide variants in type 1 and 2 long-QT syndrome. Circ. Cardiovasc. Genet. 5, 519–528 (2012).
Andreasen, C. et al. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur. J. Hum. Genet. http://dx.doi.org/10.1038/ejhg.2012.283.
Refsgaard, L. et al. High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur. J. Hum. Genet. 20, 905–908 (2012).
Ingles, J. et al. The emerging role of the cardiac genetic counselor. Heart Rhythm 8, 1958–1962 (2011).
Hershberger, R. E. et al. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ. Cardiovasc. Genet. 3, 155–161 (2010).
Ohno, S. et al. KCNE5 (KCNE1L) variants are novel modulators of Brugada syndrome and idiopathic ventricular fibrillation. Circ. Arrhythm. Electrophysiol. 4, 352–361 (2011).
Bartos, D. C. et al. A KCNQ1 mutation causes a high penetrance for familial atrial fibrillation. J. Cardiovasc. Electrophysiol. 24, 562–569 (2012).
Yang, Y. et al. Identification of a Kir3.4 mutation in congenital long QT syndrome. Am. J. Hum. Genet. 86, 872–880 (2010).
Medeiros-Domingo, A. et al. Gain-of-function mutation S422L in the KCNJ8-encoded cardiac KATP channel Kir6.1 as a pathogenic substrate for J-wave syndromes. Heart Rhythm 7, 1466–1471 (2010).
Bienengraeber, M. et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat. Genet. 36, 382–387 (2004).
Hayashi, T. et al. Identification and functional analysis of a caveolin-3 mutation associated with familial hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 313, 178–184 (2004).
Ishikawa, T. et al. A novel disease gene for Brugada syndrome: sarcolemmal membrane-associated protein gene mutations impair intracellular trafficking of hNav1.5. Circ. Arrhythm. Electrophysiol. 5, 1098–1107 (2012).
Koop, A., Goldmann, P., Chen, S. R. W., Thieleczek, R. & Varsányi, M. ARVC-related mutations in divergent region 3 alter functional properties of the cardiac ryanodine receptor. Biophys. J. 94, 4668–4677 (2008).
Crotti, L. et al. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation 127, 1009–1017 (2013).
Landstrom, A. P. et al. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J. Mol. Cell. Cardiol. 42, 1026–1035 (2007).
Landstrom, A. P., Adekola, B. A., Bos, J. M., Ommen, S. R. & Ackerman, M. J. PLN-encoded phospholamban mutation in a large cohort of hypertrophic cardiomyopathy cases: summary of the literature and implications for genetic testing. Am. Heart J. 161, 165–171 (2011).
van der Zwaag, P. A. et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur. J. Heart Fail. 14, 1199–1207 (2012).
Li, D. et al. Mutations of presenilin genes in dilated cardiomyopathy and heart failure. Am. J. Hum. Genet. 79, 1030–1039 (2006).
Landstrom, A. P. & Ackerman, M. J. Beyond the cardiac myofilament: hypertrophic cardiomyopathy-associated mutations in genes that encode calcium-handling proteins. Curr. Mol. Med. 12, 507–518 (2012).
Liu, H. et al. Molecular genetics and functional anomalies in a series of 248 Brugada cases with 11 mutations in the TRPM4 channel. PLoS ONE 8, e54131 (2013).
Ueda, K. et al. Role of HCN4 channel in preventing ventricular arrhythmia. J. Hum. Genet. 54, 115–121 (2009).
Perrot, A. et al. Prevalence of cardiac β-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J. Mol. Med. (Berl.) 83, 468–477 (2005).
Kaski, J. P. et al. Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circ. Cardiovasc. Genet. 2, 436–441 (2009).
Villard, E. et al. Mutation screening in dilated cardiomyopathy: prominent role of the β myosin heavy chain gene. Eur. Heart J. 26, 794–803 (2005).
Carniel, E. et al. α-Myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation 112, 54–59 (2005).
Karibe, A. et al. Hypertrophic cardiomyopathy caused by a novel α-tropomyosin mutation (V95A) is associated with mild cardiac phenotype, abnormal calcium binding to troponin, abnormal myosin cycling, and poor prognosis. Circulation 103, 65–71 (2001).
Watkins, H. et al. A de novo mutation in α-tropomyosin that causes hypertrophic cardiomyopathy. Circulation 91, 2302–2305 (1995).
Vasile, V. C. et al. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol. Genet. Metab. 87, 169–174 (2006).
Geier, C. et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum. Mol. Genet. 17, 2753–2765 (2008).
Hayashi, T. et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J. Am. Coll. Cardiol. 44, 2192–2201 (2004).
Duboscq-Bidot, L. et al. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur. Heart J. 30, 2128–2136 (2009).
Duboscq-Bidot, L. et al. Mutations in the Z-band protein myopalladin gene and idiopathic dilated cardiomyopathy. Cardiovasc. Res. 77, 118–125 (2008).
Mohapatra, B. et al. Mutations in the muscle LIM protein and α-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol. Genet. Metab. 80, 207–215 (2003).
Chiu, C. et al. Mutations in α-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J. Am. Coll. Cardiol. 55, 1127–1135 (2010).
Osio, A. et al. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ. Res. 100, 766–768 (2007).
Ruggiero, A., Chen, S. N., Lombardi, R., Rodriguez, G. & Marian, A. J. Pathogenesis of hypertrophic cardiomyopathy caused by myozenin 2 mutations is independent of calcineurin activity. Cardiovasc. Res. 97, 44–54 (2013).
Hassel, D. et al. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat. Med. 15, 1281–1288 (2009).
Wang, H. et al. Mutations in NEXN, a Z-disc gene, are associated with hypertrophic cardiomyopathy. Am. J. Hum. Genet. 87, 687–693 (2010).
Pilotto, A. et al. αB-crystallin mutation in dilated cardiomyopathies: low prevalence in a consecutive series of 200 unrelated probands. Biochem. Biophys. Res. Comm. 346, 1115–1117 (2006).
Sriram, C. S., Bos, J. M., Ommen, S. R. & Ackerman, M. J. Mutational analysis of CRYAB-encoded crystallin αB in hypertrophic cardiomyopathy [abstract 1625]. Circulation 116 (Suppl. II) 340 (2007).
Ferlini, A., Sewry, C., Melis, M. A., Mateddu, A. & Muntoni, F. X-linked dilated cardiomyopathy and the dystrophin gene. Neuromuscul. Disord. 9, 339–346 (1999).
Tse, H.-F. et al. Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Hum. Mol. Genet 22, 1395–1403 (2013).
Arola, A. M. et al. Mutations in PDLIM3 and MYOZ1 encoding myocyte Z line proteins are infrequently found in idiopathic dilated cardiomyopathy. Mol. Genet. Metab. 90, 435–440 (2007).
Manouvrier, S. et al. Point mutation of the mitochondrial tRNA(Leu) gene (A 3243 G) in maternally inherited hypertrophic cardiomyopathy, diabetes mellitus, renal failure, and sensorineural deafness. J. Med. Genet. 32, 654–656 (1995).
Müller, T., Krasnianski, M., Witthaut, R., Deschauer, M. & Zierz, S. Dilated cardiomyopathy may be an early sign of the C826A Fukutin-related protein mutation. Neuromuscul. Disord. 15, 372–376 (2005).
Knöll, R. et al. Laminin-α4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation 116, 515–525 (2007).
Arimura, T. et al. Mutational analysis of fukutin gene in dilated cardiomyopathy and hypertrophic cardiomyopathy. Circ. J. 73, 158–161 (2009).
Payne, R. M. & Wagner, G. R. Cardiomyopathy in Friedreich ataxia: clinical findings and research. J. Child Neurol. 27, 1179–1186 (2012).
Tsubata, S. et al. Mutations in the human δ-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J. Clin. Invest. 106, 655–662 (2000).
Erhardt, A. et al. HFE mutations in idiopathic dilated cardiomyopathy. Med. Klin. (Munich) 101 (Suppl. 1), 135–138 (2006).
Hodgkinson, K. et al. The natural history of a genetic subtype of arrhythmogenic right ventricular cardiomyopathy caused by a p.S358L mutation in TMEM43. Clin. Genet. 83, 321–331 (2013).
Maron, B. J. et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA 301, 1253–1259 (2009).
Havndrup, O. et al. Fabry disease mimicking hypertrophic cardiomyopathy: genetic screening needed for establishing the diagnosis in women. Eur. J. Heart Fail. 12, 535–540 (2010).
Kelly, B. P., Russell, M. W., Hennessy, J. R. & Ensing, G. J. Severe hypertrophic cardiomyopathy in an infant with a novel PRKAG2 gene mutation: potential differences between infantile and adult onset presentation. Pediat. Cardiol. 30, 1176–1179 (2009).
Carcavilla, A. et al. LEOPARD syndrome: a variant of Noonan syndrome strongly associated with hypertrophic cardiomyopathy. Rev. Esp. Cardiol. 6, 350–356 (2013).
Pandit, B. et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 39, 1007–1012 (2007).
Davis, J. S. et al. The overall pattern of cardiac contraction depends on a spatial gradient of myosin regulatory light chain phosphorylation. Cell 107, 631–641 (2001).
Hershberger, R. E. et al. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. J. Card. Fail. 15, 83–97 (2009).
Norgett, E. E. et al. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum. Mol. Genet. 9, 2761–2766 (2000).
Elliott, P. et al. Prevalence of desmosomal protein gene mutations in patients with dilated cardiomyopathy. Circ. Cardiovasc. Genet. 3, 314–322 (2010).
Friedrich, F. W. et al. Evidence for FHL1 as a novel disease gene for isolated hypertrophic cardiomyopathy. Hum. Mol. Genet. 21, 3237–3254 (2012).
Binder, J. S. et al. Spongious hypertrophic cardiomyopathy in patients with mutations in the four-and-a-half LIM domain 1 gene. Circ. Cardiovasc. Genet. 5, 490–502 (2012).
Arimura, T. et al. Structural analysis of four and half LIM protein-2 in dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 357, 162–167 (2007).
Beffagna, G. et al. Missense mutations in desmocollin-2 N-terminus, associated with arrhythmogenic right ventricular cardiomyopathy, affect intracellular localization of desmocollin-2 in vitro. BMC Med. Genet. 8, 65 (2007).
Bione, S. et al. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet. 12, 385–389 (1996).
Arbustini, E. et al. Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. Am. J. Pathol. 153, 1501–1510 (1998).
Levitas, A. et al. Familial neonatal isolated cardiomyopathy caused by a mutation in the flavoprotein subunit of succinate dehydrogenase. Eur. J. Hum. Genet. 18, 1160–1165 (2010).
Guo, W. et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 18, 766–773 (2012).
Quarta, G. et al. Mutations in the lamin A/C gene mimic arrhythmogenic right ventricular cardiomyopathy. Eur. Heart J. 33, 1128–1136 (2012).
Taylor, M. R. G. et al. Thymopoietin (lamina-associated polypeptide 2) gene mutation associated with dilated cardiomyopathy. Hum. Mut. 26, 566–574 (2005).
Theis, J. L. et al. Homozygosity mapping and exome sequencing reveal GATAD1 mutation in autosomal recessive dilated cardiomyopathy. Circ. Cardiovasc. Genet. 4, 585–594 (2011).
Lefeber, D. J. et al. Autosomal recessive dilated cardiomyopathy due to DOLK mutations results from abnormal dystroglycan O-mannosylation. PLoS Genet. 7, e1002427 (2011).
Beffagna, G. et al. Regulatory mutations in transforming growth factor-β3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc. Res. 65, 366–373 (2005).
Acknowledgements
A. A. M. Wilde is also affiliated with the Princess Al-Jawhara Al-Brahim Centre of Excellence in Research of Hereditary Disorders, Jeddah, Kingdom of Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article, discussed its contents, wrote the manuscript, and reviewed/edited the article before submission.
Corresponding author
Ethics declarations
Competing interests
A. A. M. Wilde declares that he is a member of the advisory board of Sorin. E. R. Behr declares no competing interests.
Rights and permissions
About this article
Cite this article
Wilde, A., Behr, E. Genetic testing for inherited cardiac disease. Nat Rev Cardiol 10, 571–583 (2013). https://doi.org/10.1038/nrcardio.2013.108
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrcardio.2013.108
This article is cited by
-
Whole exome sequencing in Brugada and long QT syndromes revealed novel rare and potential pathogenic mutations related to the dysfunction of the cardiac sodium channel
Orphanet Journal of Rare Diseases (2022)
-
A tailored approach to informing relatives at risk of inherited cardiac conditions: results of a randomised controlled trial
European Journal of Human Genetics (2022)
-
Personalized medicine in cardiovascular disease: review of literature
Journal of Diabetes & Metabolic Disorders (2021)
-
Postmortale molekulargenetische Untersuchungen (molekulare Autopsie) bei kardiovaskulären und bei ungeklärten Todesfällen
Der Kardiologe (2021)