Abstract
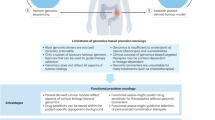
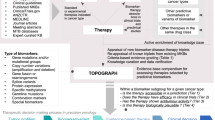
Precision medicine is about matching the right drugs to the right patients. Although this approach is technology agnostic, in cancer there is a tendency to make precision medicine synonymous with genomics. However, genome-based cancer therapeutic matching is limited by incomplete biological understanding of the relationship between phenotype and cancer genotype. This limitation can be addressed by functional testing of live patient tumour cells exposed to potential therapies. Recently, several 'next-generation' functional diagnostic technologies have been reported, including novel methods for tumour manipulation, molecularly precise assays of tumour responses and device-based in situ approaches; these address the limitations of the older generation of chemosensitivity tests. The promise of these new technologies suggests a future diagnostic strategy that integrates functional testing with next-generation sequencing and immunoprofiling to precisely match combination therapies to individual cancer patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
10 November 2015
The sentence in Box 1 referring to non-synonymous mutations should have referred to synonymous mutations. This has been corrected to state "Even synonymous mutations can have effects on cellular fitness100."
References
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Sawyers, C. L. et al. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a Phase II study. Blood 99, 3530–3539 (2002).
Talpaz, M. et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a Phase 2 study. Blood 99, 1928–1937 (2002).
Dienstmann, R., Jang, I. S., Bot, B., Friend, S. & Guinney, J. Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov. 5, 118–123 (2015).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013).
Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science 348, 56–61 (2015).
Rosenberg, S. A. & Restifo, N. P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015).
Jorgensen, J. H. & Ferraro, M. J. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49, 1749–1755 (2009).
Dishing out cancer treatment [Editorial]. Nat. Biotechnol. 31, 85 (2013).
Vidal, M., Cusick, M. E. & Barabasi, A. L. Interactome networks and human disease. Cell 144, 986–998 (2011).
Barabasi, A. L. & Oltvai, Z. N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5, 101–113 (2004).
Rolland, T. et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Piotrowska, Z. et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third generation EGFR inhibitor. Cancer Discov. 5, 713–722 (2015).
Van Allen, E. M. et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 (2014).
Johannessen, C. M. et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504, 138–142 (2013).
Burstein, H. J. et al. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 29, 3328–3330 (2011).
Samson, D. J., Seidenfeld, J., Ziegler, K. & Aronson, N. Chemotherapy sensitivity and resistance assays: a systematic review. J. Clin. Oncol. 22, 3618–3630 (2004).
Schrag, D. et al. American Society of Clinical Oncology Technology Assessment: chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 22, 3631–3638 (2004).
Von Hoff, D. D. et al. A Southwest Oncology Group study on the use of a human tumor cloning assay for predicting response in patients with ovarian cancer. Cancer 67, 20–27 (1991).
Nagourney, R. A., Evans, S. S., Messenger, J. C., Su, Y. Z. & Weisenthal, L. M. 2 chlorodeoxyadenosine activity and cross resistance patterns in primary cultures of human hematologic neoplasms. Br. J. Cancer. 67, 10–14 (1993).
Kern, D. H. & Weisenthal, L. M. Highly specific prediction of antineoplastic drug resistance with an in vitro assay using suprapharmacologic drug exposures. J. Natl Cancer Inst. 82, 582–588 (1990).
Grendys, E. C. Jr et al. Overview of a chemoresponse assay in ovarian cancer. Clin. Transl Oncol. 16, 761–769 (2014).
Maenpaa, J. U. et al. The subrenal capsule assay in selecting chemotherapy for ovarian cancer: a prospective randomized trial. Gynecol. Oncol. 57, 294–298 (1995).
Cree, I. A. et al. A prospective randomized controlled trial of tumour chemosensitivity assay directed chemotherapy versus physician's choice in patients with recurrent platinum-resistant ovarian cancer. Anticancer Drugs 18, 1093–1101 (2007).
Sandberg, R. & Ernberg, I. The molecular portrait of in vitro growth by meta-analysis of gene-expression profiles. Genome Biol. 6, R65 (2005).
Dairkee, S. H. et al. A molecular 'signature' of primary breast cancer cultures; patterns resembling tumor tissue. BMC Genomics 5, 47 (2004).
Liu, X. et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 180, 599–607 (2012).
Suprynowicz, F. A. et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl Acad. Sci. USA 109, 20035–20040 (2012).
Yuan, H. et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N. Engl. J. Med. 367, 1220–1227 (2012).
Crystal, A. S. et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 346, 1480–1486 (2014).
Baccelli, I. et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31, 539–544 (2013).
Hodgkinson, C. L. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014).
Zhang, L. et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl Med. 5, 180ra48 (2013).
Yu, M. et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345, 216–220 (2014).
Brouzes, E. et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl Acad. Sci. USA 106, 14195–14200 (2009).
Sachs, N. & Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 24, 68–73 (2014).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762–1772 (2011).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Mengelbier, L. H. et al. Intratumoral genome diversity parallels progression and predicts outcome in pediatric cancer. Nat. Commun. 6, 6125 (2015).
Junttila, M. R. & de Sauvage, F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013).
Ridky, T. W., Chow, J. M., Wong, D. J. & Khavari, P. A. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat. Med. 16, 1450–1455 (2010).
Kenny, H. A. et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat. Commun. 6, 6220 (2015).
Vaira, V. et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl Acad. Sci. USA 107, 8352–8356 (2010).
Nagourney, R. A. et al. Functional profiling to select chemotherapy in untreated, advanced or metastatic non-small cell lung cancer. Anticancer Res. 32, 4453–4460 (2012).
Majumder, B. et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun. 6, 6169 (2015).
Hirt, C. et al. 'In vitro' 3D models of tumor-immune system interaction. Adv. Drug Deliv. Rev. 79–80, 145–154 (2014).
Klinghoffer, R. A. et al. A technology platform to assess multiple cancer agents simultaneously within a patient's tumor. Sci. Transl Med. 7, 284ra58 (2015).
Jonas, O. et al. An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci. Transl Med. 7, 284ra57 (2015).
Siolas, D. & Hannon, G. J. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 73, 5315–5319 (2013).
Aparicio, S., Hidalgo, M. & Kung, A. L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer 15, 311–316 (2015).
Hidalgo, M. et al. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol. Cancer Ther. 10, 1311–1316 (2011).
Stebbing, J. et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer 120, 2006–2015 (2014).
Rubio-Viqueira, B. et al. An in vivo platform for translational drug development in pancreatic cancer. Clin. Cancer Res. 12, 4652–4661 (2006).
Eirew, P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015).
Tyner, J. W. et al. RNAi screen for rapid therapeutic target identification in leukemia patients. Proc. Natl Acad. Sci. USA 106, 8695–8700 (2009).
Tyner, J. W. et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 73, 285–296 (2013).
Glover, J. M., Loriaux, M., Tyner, J. W., Druker, B. J. & Chang, B. H. In vitro sensitivity to dasatinib in lymphoblasts from a patient with t(17;19)(q22;p13) gene rearrangement pre-B acute lymphoblastic leukemia. Pediatr. Blood Cancer 59, 576–579 (2012).
Kulesz-Martin, M. F. et al. A molecular case report: functional assay of tyrosine kinase inhibitors in cells from a patient's primary renal cell carcinoma. Cancer Biol. Ther. 14, 95–99 (2013).
Davis, L. E. et al. A case study of personalized therapy for osteosarcoma. Pediatr. Blood Cancer 60, 1313–1319 (2013).
Pemovska, T. et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 3, 1416–1429 (2013).
Yadav, B. et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci. Rep. 4, 5193 (2014).
Pemovska, T. et al. Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature 519, 102–105 (2015).
Klco, J. M. et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood 121, 1633–1643 (2013).
Lamothe, B. et al. Proteasome inhibitor carfilzomib complements ibrutinib's action in chronic lymphocytic leukemia. Blood 125, 407–410 (2015).
Hilhorst, R. et al. Peptide microarrays for profiling of serine/threonine kinase activity of recombinant kinases and lysates of cells and tissue samples. Methods Mol. Biol. 977, 259–271 (2013).
Tahiri, A. et al. Differential inhibition of ex-vivo tumor kinase activity by vemurafenib in BRAF(V600E) and BRAF wild-type metastatic malignant melanoma. PLoS ONE 8, e72692 (2013).
Schayowitz, A. et al. Functional profiling of live melanoma samples using a novel automated platform. PLoS ONE 7, e52760 (2012).
Irish, J. M. et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell 118, 217–228 (2004).
Krutzik, P. O., Crane, J. M., Clutter, M. R. & Nolan, G. P. High-content single-cell drug screening with phosphospecific flow cytometry. Nat. Chem. Biol. 4, 132–142 (2008).
Kornblau, S. M. et al. Dynamic single-cell network profiles in acute myelogenous leukemia are associated with patient response to standard induction therapy. Clin. Cancer Res. 16, 3721–3733 (2010).
Lacayo, N. J. et al. Development and validation of a single-cell network profiling assay-based classifier to predict response to induction therapy in paediatric patients with de novo acute myeloid leukaemia: a report from the Children's Oncology Group. Br. J. Haematol. 162, 250–262 (2013).
Hata, A. N., Engelman, J. A. & Faber, A. C. The BCL2 family: key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Discov. 5, 475–487 (2015).
Del Gaizo Moore, V. & Letai, A. BH3 profiling — measuring integrated function of the mitochondrial apoptotic pathway to predict cell fate decisions. Cancer Lett. 332, 202–205 (2013).
Ni Chonghaile, T. et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science 334, 1129–1133 (2011).
Vo, T. T. et al. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell 151, 344–355 (2012).
Montero, J. et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell 160, 977–989 (2015).
Jahnke, H. G. et al. Direct chemosensitivity monitoring ex vivo on undissociated melanoma tumor tissue by impedance spectroscopy. Cancer Res. 74, 6408–6418 (2014).
Kleinhans, R. et al. Sensor-based cell and tissue screening for personalized cancer chemotherapy. Med. Biol. Eng. Comput. 50, 117–126 (2012).
Simon, R. & Roychowdhury, S. Implementing personalized cancer genomics in clinical trials. Nat. Rev. Drug Discov. 12, 358–369 (2013).
Sawyers, C. L. & van 't Veer, L. J. Reliable and effective diagnostics are keys to accelerating personalized cancer medicine and transforming cancer care: a policy statement from the American Association for Cancer Research. Clin. Cancer Res. 20, 4978–4981 (2014).
U.S. Department of Health and Human Services. Anticipated Details of the Draft Guidance to Industry, Food and Drug Administration Staff and Clinical Laboratories: Framework for Regulatory Oversight of Laboratory Developed Tests (LDTs) FDA [online], (2014).
Kreiter, S. et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696 (2015).
Wheler, J. J. et al. Prospective study comparing outcomes in patients with advanced malignancies on molecular alteration-matched versus non-matched therapy. J. Clin. Oncol. 33, S11019 (2015).
Chapman, P. B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011).
Flaherty, K. T. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 (2010).
Mok, T. S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361, 947–957 (2009).
Kwak, E. L. et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 363, 1693–1703 (2010).
Holohan, C., Van Schaeybroeck, S., Longley, D. B. & Johnston, P. G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13, 714–726 (2013).
Wander, S. A., Levis, M. J. & Fathi, A. T. The evolving role of FLT3 inhibitors in acute myeloid leukemia: quizartinib and beyond. Ther. Adv. Hematol. 5, 65–77 (2014).
Kopetz, S. et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J. Clin. Oncol. 28, 15s (2010).
Rubio-Perez, C. et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 27, 382–396 (2015).
Herbst, R. S. et al. Lung Master Protocol (Lung-MAP) — a biomarker-driven protocol for accelerating development of therapies for squamous cell lung cancer: SWOG S1400. Clin. Cancer Res. 21, 1514–1524 (2015).
Lopez-Chavez, A. et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology Phase II basket trial. J. Clin. Oncol. 33, 1000–1007 (2015).
Kris, M. G. et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 311, 1998–2006 (2014).
Lind, P. A., Berg, O. G. & Andersson, D. I. Mutational robustness of ribosomal protein genes. Science 330, 825–827 (2010).
Cameron, F. & Sanford, M. Ibrutinib: first global approval. Drugs 74, 263–271 (2014).
Markham, A. Idelalisib: first global approval. Drugs 74, 1701–1707 (2014).
Luo, J., Solimini, N. L. & Elledge, S. J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837 (2009).
Haibe-Kains, B. et al. Inconsistency in large pharmacogenomic studies. Nature 504, 389–393 (2013).
Hatzis, C. et al. Enhancing reproducibility in cancer drug screening: how do we move forward? Cancer Res. 74, 4016–4023 (2014).
Simon, R. M., Paik, S. & Hayes, D. F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl Cancer Inst. 101, 1446–1452 (2009).
Tsourounis, M., Stuart, J., Pignato, W., Toscani, M. & Barone, J. Current trends in personalized medicine and companion diagnostics: a summary from the DIA meeting on personalized medicine and companion diagnostics. Ther. Innov. Regul. Sci. 49, 530–543 (2015).
Acknowledgements
The authors apologize to investigators whose research they were unable to include owing to space considerations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
A.L. is a co-inventor on patents related to BH3 profiling for the selection of therapies for patients with cancer. A.A.F., A.L., D.E.F. and K.T.F. hold equity in and serve on the Scientific Advisory Board of Leap Oncology.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Friedman, A., Letai, A., Fisher, D. et al. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer 15, 747–756 (2015). https://doi.org/10.1038/nrc4015
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc4015
This article is cited by
-
Culture and expansion of murine proximal airway basal stem cells
Stem Cell Research & Therapy (2024)
-
Platform trial for off-label oncology drugs using comprehensive genomic profiling under the universal public healthcare system: the BELIEVE trial
International Journal of Clinical Oncology (2024)
-
Precision medicine applied to metastatic colorectal cancer using tumor-derived organoids and in-vitro sensitivity testing: a phase 2, single-center, open-label, and non-comparative study
Journal of Experimental & Clinical Cancer Research (2023)
-
Dual network analysis of transcriptome data for discovery of new therapeutic targets in non-small cell lung cancer
Oncogene (2023)
-
Microphysiological model reveals the promise of memory-like natural killer cell immunotherapy for HIV± cancer
Nature Communications (2023)