Abstract
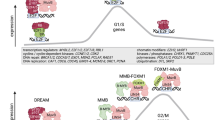
The dimerization partner, RB-like, E2F and multi-vulval class B (DREAM) complex provides a previously unsuspected unifying role in the cell cycle by directly linking p130, p107, E2F, BMYB and forkhead box protein M1. DREAM mediates gene repression during the G0 phase and coordinates periodic gene expression with peaks during the G1/S and G2/M phases. Perturbations in DREAM complex regulation shift the balance from quiescence towards proliferation and contribute to the increased mitotic gene expression levels that are frequently observed in cancers with a poor prognosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
23 August 2013
On page 593 of this Opinion article, a citation to a key reference (Flowers. S., Beck, G. R. Jr & Moran, E. Tissue-specific gene targeting by the multiprotein mammalian DREAM complex. J. Biol. Chem. 286, 27867–27871 (2011)) was not included. This reference should have been cited at the end of the following sentence: "Perhaps isolation of the DREAM and the BMYB–MuvB complexes from distinct mammalian tissues will reveal functions in addition to cell cycle control130,131". This has now been corrected online.
References
Whitfield, M. L. et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977–2000 (2002).
Ferguson, E. L., Sternberg, P. W. & Horvitz, H. R. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326, 259–267 (1987).
Aroian, R. V., Koga, M., Mendel, J. E., Ohshima, Y. & Sternberg, P. W. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 348, 693–699 (1990).
Beitel, G. J., Clark, S. G. & Horvitz, H. R. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348, 503–509 (1990).
Han, M. & Sternberg, P. W. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell 63, 921–931 (1990).
Ferguson, E. L. & Horvitz, H. R. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics 123, 109–121 (1989).
Harrison, M. M., Lu, X. & Horvitz, H. R. LIN-61, one of two Caenorhabditis elegans malignant-brain-tumor-repeat-containing proteins, acts with the DRM and NuRD-like protein complexes in vulval development but not in certain other biological processes. Genetics 176, 255–271 (2007).
Ceol, C. J. & Horvitz, H. R. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol. Cell 7, 461–473 (2001).
Fay, D. S. & Han, M. The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis 26, 279–284 (2000).
Beitel, G. J., Lambie, E. J. & Horvitz, H. R. The C. elegans gene lin-9, which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein. Gene 254, 253–263 (2000).
Thomas, J. H., Ceol, C. J., Schwartz, H. T. & Horvitz, H. R. New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans. Genetics 164, 135–151 (2003).
McMahon, S. B., Van Buskirk, H. A., Dugan, K. A., Copeland, T. D. & Cole, M. D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94, 363–374 (1998).
Ceol, C. J. & Horvitz, H. R. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6, 563–576 (2004).
Calvi, B. R. & Spradling, A. C. Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods 18, 407–417 (1999).
Beall, E. L. et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420, 833–837 (2002).
Lewis, P. W. et al. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 18, 2929–2940 (2004).
Beall, E. L., Bell, M., Georlette, D. & Botchan, M. R. Dm-myb mutant lethality in Drosophila is dependent upon mip130: positive and negative regulation of DNA replication. Genes Dev. 18, 1667–1680 (2004).
Korenjak, M. et al. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119, 181–193 (2004).
Dimova, D. K., Stevaux, O., Frolov, M. V. & Dyson, N. J. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17, 2308–2320 (2003).
Taylor-Harding, B., Binne, U. K., Korenjak, M., Brehm, A. & Dyson, N. J. p55, the Drosophila ortholog of RbAp46/RbAp48, is required for the repression of dE2F2/RBF-regulated genes. Mol. Cell. Biol. 24, 9124–9136 (2004).
Harrison, M. M., Ceol, C. J., Lu, X. & Horvitz, H. R. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl Acad. Sci. USA 103, 16782–16787 (2006).
Davidson, C. J., Guthrie, E. E. & Lipsick, J. S. Duplication and maintenance of the Myb genes of vertebrate animals. Biol. Open 2, 101–110 (2013).
Gagrica, S. et al. Inhibition of oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J. 23, 4627–4638 (2004).
Sandoval, R. et al. A mutant allele of BARA/LIN-9 rescues the cdk4−/− phenotype by releasing the repression on E2F-regulated genes. Exp. Cell Res. 312, 2465–2475 (2006).
Litovchick, L. et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26, 539–551 (2007).
Schmit, F. et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle 6, 1903–1913 (2007).
Pilkinton, M., Sandoval, R. & Colamonici, O. R. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene 26, 7535–7543 (2007).
Joaquin, M., Bessa, M., Saville, M. K. & Watson, R. J. B-Myb overcomes a p107-mediated cell proliferation block by interacting with an N-terminal domain of p107. Oncogene 21, 7923–7932 (2002).
Chicas, A. et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17, 376–387 (2010).
Litovchick, L., Florens, L. A., Swanson, S. K., Washburn, M. P. & DeCaprio, J. A. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 25, 801–813 (2011).
Takahashi, Y., Rayman, J. B. & Dynlacht, B. D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14, 804–816 (2000).
Tedesco, D., Lukas, J. & Reed, S. I. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 16, 2946–2957 (2002).
Georlette, D. et al. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 21, 2880–2896 (2007).
Hurford, R. K. Jr., Cobrinik, D., Lee, M. H. & Dyson, N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11, 1447–1463 (1997).
Reichert, N. et al. Lin9, a subunit of the mammalian DREAM complex, is essential for embryonic development, for survival of adult mice, and for tumor suppression. Mol. Cell. Biol. 30, 2896–2908 (2010).
Gaubatz, S. et al. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6, 729–735 (2000).
Zhu, W., Giangrande, P. H. & Nevins, J. R. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 23, 4615–4626 (2004).
Muller, G. A. & Engeland, K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 277, 877–893 (2010).
Muller, G. A. et al. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 40, 1561–1578 (2012).
Schmit, F., Cremer, S. & Gaubatz, S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 276, 5703–5716 (2009).
Catchpole, S., Tavner, F., Le Cam, L., Sardet, C. & Watson, R. J. A. B-myb promoter corepressor site facilitates in vivo occupation of the adjacent E2F site by p107 x E2F and p130 x E2F complexes. J. Biol. Chem. 277, 39015–39024 (2002).
Nakajima, Y., Yamada, S., Kamata, N. & Ikeda, M. A. Interaction of E2F-Rb family members with corepressors binding to the adjacent E2F site. Biochem. Biophys. Res. Commun. 364, 1050–1055 (2007).
Aranda, S., Laguna, A. & de la Luna, S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 25, 449–462 (2011).
Tschop, K. et al. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 25, 814–830 (2011).
Zhao, B., Li, L., Tumaneng, K., Wang, C. Y. & Guan, K. L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev. 24, 72–85 (2010).
Wang, K., Degerny, C., Xu, M. & Yang, X. J. YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem. Cell Biol. 87, 77–91 (2009).
Harvey, K. F., Zhang, X. & Thomas, D. M. The Hippo pathway and human cancer. Nature Rev. Cancer 13, 246–257 (2013).
Huang, J., Wu, S., Barrera, J., Matthews, K. & Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434 (2005).
Zhao, B. et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007).
Hao, Y., Chun, A., Cheung, K., Rashidi, B. & Yang, X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 (2008).
Nishioka, N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009).
Sandoval, R., Pilkinton, M. & Colamonici, O. R. Deletion of the p107/p130-binding domain of Mip130/LIN-9 bypasses the requirement for CDK4 activity for the dissociation of Mip130/LIN-9 from p107/p130-E2F4 complex. Exp. Cell Res. 315, 2914–2920 (2009).
Nor Rashid, N., Yusof, R. & Watson, R. J. Disruption of repressive p130-DREAM complexes by human papillomavirus 16 E6/E7 oncoproteins is required for cell-cycle progression in cervical cancer cells. J. Gen. Virol. 92, 2620–2627 (2011).
Sadasivam, S., Duan, S. & DeCaprio, J. A. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 26, 474–489 (2012).
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Collado, M. & Serrano, M. Senescence in tumours: evidence from mice and humans. Nature Rev. Cancer 10, 51–57 (2010).
Kosar, M. et al. Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16ink4a. Cell Cycle 10, 457–468 (2011).
Knight, A. S., Notaridou, M. & Watson, R. J. A. Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene 28, 1737–1747 (2009).
Lam, E. W., Robinson, C. & Watson, R. J. Characterization and cell cycle-regulated expression of mouse B-myb. Oncogene 7, 1885–1890 (1992).
Pilkinton, M., Sandoval, R., Song, J., Ness, S. A. & Colamonici, O. R. Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1. J. Biol. Chem. 282, 168–175 (2007).
Sala, A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur. J. Cancer 41, 2479–2484 (2005).
Joaquin, M. & Watson, R. J. Cell cycle regulation by the B-Myb transcription factor. Cell. Mol. Life Sci. 60, 2389–2401 (2003).
Zhan, M. et al. The B-MYB transcriptional network guides cell cycle progression and fate decisions to sustain self-renewal and the identity of pluripotent stem cells. PLoS ONE 7, e42350 (2012).
Ansieau, S., Kowenz-Leutz, E., Dechend, R. & Leutz, A. B-Myb, a repressed trans-activating protein. J. Mol. Med. 75, 815–819 (1997).
Lane, S., Farlie, P. & Watson, R. B-Myb function can be markedly enhanced by cyclin A-dependent kinase and protein truncation. Oncogene 14, 2445–2453 (1997).
Sala, A. et al. Activation of human B-MYB by cyclins. Proc. Natl Acad. Sci. USA 94, 532–536 (1997).
Ziebold, U., Bartsch, O., Marais, R., Ferrari, S. & Klempnauer, K. H. Phosphorylation and activation of B-Myb by cyclin A-Cdk2. Curr. Biol. 7, 253–260 (1997).
Johnson, T. K., Schweppe, R. E., Septer, J. & Lewis, R. E. Phosphorylation of B-Myb regulates its transactivation potential and DNA binding. J. Biol. Chem. 274, 36741–36749 (1999).
Saville, M. K. & Watson, R. J. The cell-cycle regulated transcription factor B-Myb is phosphorylated by cyclin A/Cdk2 at sites that enhance its transactivation properties. Oncogene 17, 2679–2689 (1998).
Katzen, A. L. et al. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev. 12, 831–843 (1998).
Okada, M., Akimaru, H., Hou, D. X., Takahashi, T. & Ishii, S. Myb controls G(2)/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J. 21, 675–684 (2002).
Wen, H., Andrejka, L., Ashton, J., Karess, R. & Lipsick, J. S. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 22, 601–614 (2008).
Fung, S. M., Ramsay, G. & Katzen, A. L. Mutations in Drosophila myb lead to centrosome amplification and genomic instability. Development 129, 347–359 (2002).
Manak, J. R., Mitiku, N. & Lipsick, J. S. Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc. Natl Acad. Sci. USA 99, 7438–7443 (2002).
Shepard, J. L. et al. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc. Natl Acad. Sci. USA 102, 13194–13199 (2005).
Osterloh, L. et al. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J. 26, 144–157 (2007).
Beall, E. L. et al. Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 21, 904–919 (2007).
Davidson, C. J., Tirouvanziam, R., Herzenberg, L. A. & Lipsick, J. S. Functional evolution of the vertebrate Myb gene family: B-Myb, but neither A-Myb nor c-Myb, complements Drosophila Myb in hemocytes. Genetics 169, 215–229 (2005).
Kittler, R. et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nature Cell Biol. 9, 1401–1412 (2007).
Matsuo, T., Kuramoto, H., Kumazaki, T., Mitsui, Y. & Takahashi, T. LIN54 harboring a mutation in CHC domain is localized to the cytoplasm and inhibits cell cycle progression. Cell Cycle 11, 3227–3236 (2012).
Charrasse, S., Carena, I., Brondani, V., Klempnauer, K. H. & Ferrari, S. Degradation of B-Myb by ubiquitin-mediated proteolysis: involvement of the Cdc34-SCF(p45Skp2) pathway. Oncogene 19, 2986–2995 (2000).
Down, C. F., Millour, J., Lam, E. W. & Watson, R. J. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim. Biophys. Acta 1819, 855–862 (2012).
Chen, X. et al. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell. Biol. 33, 227–236 (2013).
Laoukili, J. et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nature Cell Biol. 7, 126–136 (2005).
Wang, I. C. et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25, 10875–10894 (2005).
Korver, W., Roose, J., Wilson, A. & Clevers, H. The winged-helix transcription factor Trident is expressed in actively dividing lymphocytes. Immunobiology 198, 157–161 (1997).
Laoukili, J. et al. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol. Cell. Biol. 28, 3076–3087 (2008).
Park, H. J., Costa, R. H., Lau, L. F., Tyner, A. L. & Raychaudhuri, P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol. Cell. Biol. 28, 5162–5171 (2008).
Korver, W. et al. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor Trident. Curr. Biol. 8, 1327–1330 (1998).
Lefebvre, C. et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol. 6, 377 (2010).
Zhou, Y. & Ness, S. A. Myb proteins: angels and demons in normal and transformed cells. Front. Biosci. 16, 1109–1131 (2011).
Lorvellec, M. et al. B-Myb is critical for proper DNA duplication during an unperturbed S phase in mouse embryonic stem cells. Stem Cells 28, 1751–1759 (2010).
Littler, D. R. et al. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 38, 4527–4538 (2010).
Fu, Z. et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nature Cell Biol. 10, 1076–1082 (2008).
Thomas, L. R. & Tansey, W. P. Proteolytic control of the oncoprotein transcription factor Myc. Adv. Cancer Res. 110, 77–106 (2011).
Whitfield, M. L., George, L. K., Grant, G. D. & Perou, C. M. Common markers of proliferation. Nature Rev. Cancer 6, 99–106 (2006).
Rhodes, D. R. et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl Acad. Sci. USA 101, 9309–9314 (2004).
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Thorner, A. R. et al. In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene 28, 742–751 (2009).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
O'Connell, M. J. et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J. Clin. Oncol. 28, 3937–3944 (2010).
Tian, S. et al. Biological functions of the genes in the mammaprint breast cancer profile reflect the hallmarks of cancer. Biomark. Insights 5, 129–138 (2010).
Garcia, P., Berlanga, O., Watson, R. & Frampton, J. Generation of a conditional allele of the B-myb gene. Genesis 43, 189–195 (2005).
Clarke, M. et al. MYBL2 haploinsufficiency increases susceptibility to age-related haematopoietic neoplasia. Leukemia 27, 661–670 (2013).
Testa, J. R., Kinnealey, A., Rowley, J. D., Golde, D. W. & Potter, D. Deletion of the long arm of chromosome 20 [del(20)(q11)] in myeloid disorders. Blood 52, 868–877 (1978).
Mullier, F. et al. Morphology, cytogenetics, and survival in myelodysplasia with del(20q) or ider(20q): a multicenter study. Ann. Hematol. 91, 203–213 (2012).
Liu, M. et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 66, 3593–3602 (2006).
Kalin, T. V. et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 66, 1712–1720 (2006).
Kim, I. M. et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 66, 2153–2161 (2006).
Dibb, M. et al. The FOXM1-PLK1 axis is commonly upregulated in oesophageal adenocarcinoma. Br. J. Cancer 107, 1766–1775 (2012).
Wang, Z., Banerjee, S., Kong, D., Li, Y. & Sarkar, F. H. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 67, 8293–8300 (2007).
Francis, R. E. et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int. J. Oncol. 35, 57–68 (2009).
Lok, G. T. et al. Aberrant activation of ERK/FOXM1 signaling cascade triggers the cell migration/invasion in ovarian cancer cells. PLoS ONE 6, e23790 (2011).
Carter, S. L., Eklund, A. C., Kohane, I. S., Harris, L. N. & Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nature Genet. 38, 1043–1048 (2006).
Masselink, H., Vastenhouw, N. & Bernards, R. B-myb rescues ras-induced premature senescence, which requires its transactivation domain. Cancer Lett. 171, 87–101 (2001).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Hu, J., Deng, H. & Friedman, E. A. Ovarian cancer cells, not normal cells, are damaged by Mirk/Dyrk1B kinase inhibition. Int. J. Cancer 132, 2258–2269 (2013).
Mannefeld, M., Klassen, E. & Gaubatz, S. B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res. 69, 4073–4080 (2009).
Quaas, M., Muller, G. A. & Engeland, K. p53 can repress transcription of cell cycle genes through a p21WAF1/CIP1-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle 11, 4661–4672 (2012).
Toettcher, J. E. et al. Distinct mechanisms act in concert to mediate cell cycle arrest. Proc. Natl Acad. Sci. USA 106, 785–790 (2009).
Giangrande, P. H. et al. A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 18, 2941–2951 (2004).
Westendorp, B. et al. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res. 40, 3511–3523 (2012).
Christensen, J. et al. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 33, 5458–5470 (2005).
Logan, N. et al. E2F-8: an E2F family member with a similar organization of DNA-binding domains to E2F-7. Oncogene 24, 5000–5004 (2005).
Maiti, B. et al. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 280, 18211–18220 (2005).
Mohammed, H. et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3, 342–349 (2013).
Sim, C. K., Perry, S., Tharadra, S. K., Lipsick, J. S. & Ray, A. Epigenetic regulation of olfactory receptor gene expression by the Myb-MuvB/dREAM complex. Genes Dev. 26, 2483–2498 (2012).
Rovani, M. K., Brachmann, C. B., Ramsay, G. & Katzen, A. L. The dREAM/Myb-MuvB complex and Grim are key regulators of the programmed death of neural precursor cells at the Drosophila posterior wing margin. Dev. Biol. 372, 88–102 (2012).
Petrella, L. N. et al. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138, 1069–1079 (2011).
Lewis, P. W. et al. Drosophila lin-52 acts in opposition to repressive components of the Myb-MuvB/dREAM complex. Mol. Cell. Biol. 32, 3218–3227 (2012).
Tabuchi, T. M. et al. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 7, e1002074 (2011).
Swanton, C. et al. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature 390, 184–187 (1997).
Ellis, M. et al. Degradation of p27Kip cdk inhibitor triggered by Kaposi's sarcoma virus cyclin–cdk6 complex. EMBO J. 18, 644–653 (1999).
Aksoy, O. et al. The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence. Genes Dev. 26, 1546–1557 (2012).
Carvajal, L. A., Hamard, P. J., Tonnessen, C. & Manfredi, J. J. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 26, 1533–1545 (2012).
Chen, H. Z. et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nature Cell Biol. 14, 1192–1202 (2012).
Slansky, J. E. & Farnham, P. J. Introduction to the E2F family: protein structure and gene regulation. Curr. Top. Microbiol. Immunol. 208, 1–30 (1996).
Tao, Y., Kassatly, R. F., Cress, W. D. & Horowitz, J. M. Subunit composition determines E2F DNA-binding site specificity. Mol. Cell. Biol. 17, 6994–7007 (1997).
Ness, S. A., Marknell, A. & Graf, T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59, 1115–1125 (1989).
Mizuguchi, G. et al. DNA binding activity and transcriptional activator function of the human B-myb protein compared with c-MYB. J. Biol. Chem. 265, 9280–9284 (1990).
Korver, W., Roose, J. & Clevers, H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 25, 1715–1719 (1997).
Flowers, S., G. R. Jr & Moran, E. Tissue-specific gene targeting by the multiprotein mammalian DREAM complex. J. Biol. Chem. 286, 27867–27871 (2011).
Acknowledgements
The authors gratefully acknowledge the many helpful discussions about DREAM with L. Litovchick, N. Dyson, M. Botchan and K. Engeland. This work was supported in part by US Public Health Service grants P01CA050661 and R01CA63113 to J.A.D.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
DATABASES
Rights and permissions
About this article
Cite this article
Sadasivam, S., DeCaprio, J. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 13, 585–595 (2013). https://doi.org/10.1038/nrc3556
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3556
This article is cited by
-
Bioinformatic identification and experiment validation reveal 6 hub genes, promising diagnostic and therapeutic targets for Alzheimer’s disease
BMC Medical Genomics (2024)
-
Mirk/Dyrk1B controls ventral spinal cord development via Shh pathway
Cellular and Molecular Life Sciences (2024)
-
BCAS2 regulates granulosa cell survival by participating in mRNA alternative splicing
Journal of Ovarian Research (2023)
-
TRABID inhibition activates cGAS/STING-mediated anti-tumor immunity through mitosis and autophagy dysregulation
Nature Communications (2023)
-
Drug-tolerant persister cells in cancer: the cutting edges and future directions
Nature Reviews Clinical Oncology (2023)