Key Points
-
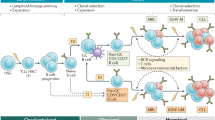
Chronic lymphocytic leukaemia (CLL) is the most common leukaemia in the Western world. It is characterized by the accumulation of small B lymphocytes that have a mature appearance.
-
Two subsets of CLL cases can be differentiated by the degree of somatic hypermutation (mutated and unmutated immunoglobulin heavy chain variable region (IGHV) genes) that have distinct clinical and biological behaviours.
-
Overall, more than 20% of CLL cases carry stereotyped B cell receptors, suggesting that common antigen(s) are recognized by CLL cells.
-
Clonal B cell populations with a CLL immunophenotype have been detected in 3.5% of healthy individuals (monoclonal B cell lymphocytosis; MBL). MBL is often a CLL precursor.
-
Approximately 80% of CLLs show aberrations in a few frequently affected chromosomal regions, including 13q14 (mir-15a and mir16-1), 11q23 (ataxia telangiectasia-mutated; ATM), trisomy 12 and 17p13 (TP53). Recurrent translocations are rare in CLL.
-
Global and gene-specific aberrant DNA methylation has been detected in CLL. Almost all sporadic CLL cases also show epigenetic silencing of death-associated protein kinase 1.
-
In lymphoid organs, CLL cells interact with and seem to shape their microenvironment, which consists of T cells, stromal cells and soluble factors. This interaction is emerging as a therapeutic target.
-
p53 plays a central part in our current understanding of why some patients fail to respond to chemotherapy.
-
The most powerful prognostic factors include 17p13 deletion, TP53 mutation, 11q23 deletion, IGHV mutation status, serum markers, clinical stage and age.
-
CLL may serve as a model of how microenvironmental stimuli, antigenic drive and epigenetic, as well as genetic, deregulation are combined in cancer pathogenesis.
Abstract
Chronic lymphocytic leukaemia (CLL) has several unique features that distinguish it from other cancers. Most CLL tumour cells are inert and arrested in G0/G1 of the cell cycle and there is only a small proliferative compartment; however, the progressive accumulation of malignant cells will ultimately lead to symptomatic disease. Pathogenic mechanisms have been elucidated that involve multiple external (for example, microenvironmental stimuli and antigenic drive) and internal (genetic and epigenetic) events that are crucial in the transformation, progression and evolution of CLL. Our growing understanding of CLL biology is allowing the translation of targets and biological classifiers into clinical practice.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dores, G. M. et al. Chronic lymphocytic leukaemia and small lymphocytic lymphoma: overview of the descriptive epidemiology. Br. J. Haematol. 139, 809–819 (2007).
Goldin, L. R., Pfeiffer, R. M., Li, X. & Hemminki, K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood 104, 1850–1854 (2004).
Hallek, M. et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 111, 5446–5456 (2008).
Chiorazzi, N., Rai, K. R. & Ferrarini, M. Chronic lymphocytic leukemia. N. Engl. J. Med. 352, 804–815 (2005).
Binet, J. L. et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer 48, 198–206 (1981).
Rai, K. R. et al. Clinical staging of chronic lymphocytic leukemia. Blood 46, 219–234 (1975).
Fais, F. et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Invest. 102, 1515–1525 (1998).
Damle, R. N. et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94, 1840–1847 (1999).
Hamblin, T. J., Davis, Z., Gardiner, A., Oscier, D. G. & Stevenson, F. K. Unmutated Ig VH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 94, 1848–1854 (1999). The two pivotal studies by Hamblin et al . and Damle et al . establish the prognostic impact of IGHV mutational status in CLL.
Tobin, G. et al. Chronic lymphocytic leukemias utilizing the VH3-21 gene display highly restricted Vλ2-14 gene use and homologous CDR3s: implicating recognition of a common antigen epitope. Blood 101, 4952–4957 (2003).
Messmer, B. T., Albesiano, E., Messmer, D. & Chiorazzi, N. The pattern and distribution of immunoglobulin VH gene mutations in chronic lymphocytic leukemia B cells are consistent with the canonical somatic hypermutation process. Blood 103, 3490–3495 (2004).
Ghia, P. et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3-21 gene. Blood 105, 1678–1685 (2005).
Stamatopoulos, K. et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood 109, 259–270 (2007).
Schroeder, H. W. Jr & Dighiero, G. The pathogenesis of chronic lymphocytic leukemia: analysis of the antibody repertoire. Immunol. Today 15, 288–294 (1994). An analysis indicating that CLL cells use a IGHV repertoire that is characteristic of mature B cells, and suggests that antigens may play a part in the pathogenesis of this disease.
Calin, G. A. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci USA 99, 15524–15529 (2002). This study establishes the link between 13q14 deletion and downregulation of miR-15a and miR-16-1 in CLL.
Cimmino, A. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA 102, 13944–13949 (2005).
Döhner, H. et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood 85, 1580–1589 (1995).
Raval, A. et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell 129, 879–890 (2007). This study describes a pathogenetic link between downregulation of DAPK1 (by methylation) and CLL.
Schaffner, C., Stilgenbauer, S., Rappold, G. A., Döhner, H. & Lichter, P. Somatic ATM mutations indicate a pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood 94, 748–753 (1999).
Austen, B. et al. Mutations in the ATM gene lead to impaired overall and treatment-free survival that is independent of IGVH mutation status in patients with B-CLL. Blood 106, 3175–3182 (2005).
Austen, B. et al. Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. J. Clin. Oncol. 25, 5448–5457 (2007).
Döhner, H. et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 343, 1910–1916 (2000). Development of the hierarchical model of CLL prognosis based on recurrent genomic aberrations.
Zenz, T. et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood 112, 3322–3329 (2008).
Klein, U. & Dalla-Favera, R. Germinal centres: role in B-cell physiology and malignancy. Nature Rev. Immunol. 8, 22–33 (2008).
Küppers, R. Mechanisms of B-cell lymphoma pathogenesis. Nature Rev. Cancer 5, 251–262 (2005).
Kröber, A. et al. VH mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood 100, 1410–1416 (2002).
Hamblin, T. J., Davis, Z. A. & Oscier, D. G. Determination of how many immunoglobulin variable region heavy chain mutations are allowable in unmutated chronic lymphocytic leukaemia — long-term follow up of patients with different percentages of mutations. Br. J. Haematol. 140, 320–323 (2008).
Gurrieri, C. et al. Chronic lymphocytic leukemia B cells can undergo somatic hypermutation and intraclonal immunoglobulin VHDJH gene diversification. J. Exp. Med. 196, 629–639 (2002).
Albesiano, E. et al. Activation-induced cytidine deaminase in chronic lymphocytic leukemia B cells: expression as multiple forms in a dynamic, variably sized fraction of the clone. Blood 102, 3333–3339 (2003).
Oscier, D. G., Thompsett, A., Zhu, D. & Stevenson, F. K. Differential rates of somatic hypermutation in VH genes among subsets of chronic lymphocytic leukemia defined by chromosomal abnormalities. Blood 89, 4153–4160 (1997).
Klein, U. et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp. Med. 194, 1625–1638 (2001). This study demonstrates that the global gene expression of IGHV-mutated and unmutated CLLs are more similar to memory B cells than to naive or CD5+ B cells.
Rosenwald, A. et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J. Exp. Med. 194, 1639–1647 (2001). This study compared gene expression profiles of CLL samples with unmutated and mutated IGHV and discovered that ZAP70 was differentially expressed in these subgroups.
Damle, R. N. et al. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood 99, 4087–4093 (2002).
34. Damle, R. N. et al. Telomere length and telomerase activity delineate distinctive replicative features of the B-CLL subgroups defined by immunoglobulin V gene mutations. Blood 103, 375–382 (2004).
Stilgenbauer, S. et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica 92, 1242–1245 (2007).
Kipps, T. J. The B-cell receptor and ZAP-70 in chronic lymphocytic leukemia. Best. Pract. Res. Clin. Haematol. 20, 415–424 (2007).
Chen, L. et al. ZAP-70 directly enhances IgM signaling in chronic lymphocytic leukemia. Blood 105, 2036–2041 (2005).
Lanham, S. et al. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood 101, 1087–1093 (2003).
Mockridge, C. I. et al. Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood 109, 4424–4431 (2007).
Guarini, A. et al. BCR-ligation induced by IgM stimulation results in gene expression and functional changes only in IgVH unmutated chronic lymphocytic leukemia (CLL) cells. Blood 112, 782–792 (2008).
Muzio, M. et al. Constitutive activation of distinct BCR-signaling pathways in a subset of CLL patients: a molecular signature of anergy. Blood 112, 188–195 (2008).
Hadzidimitriou, A. et al. Evidence for the significant role of immunoglobulin light chains in antigen recognition and selection in chronic lymphocytic leukemia. Blood 113, 403–411 (2009).
Widhopf, G. F. et al. Nonstochastic pairing of immunoglobulin heavy and light chains expressed by chronic lymphocytic leukemia B cells is predicated on the heavy chain CDR3. Blood 111, 3137–3144 (2008).
Messmer, B. T. et al. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J. Exp. Med. 200, 519–525 (2004). The demonstration of a striking degree of structural restriction of the entire BCR in CLL, which suggested that common antigens could be recognized by CLL cells.
Widhopf, G. F. et al. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood 104, 2499–2504 (2004). The findings of virtually identical Igs in 1.3% of CLLs provided compelling evidence that the Igs expressed by CLL B cells are highly selected and unlike the Igs expressed by naive B cells.
Murray, F. et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood 111, 1524–1533 (2008).
Chiorazzi, N. & Ferrarini, M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu. Rev. Immunol. 21, 841–894 (2003).
Herve, M. et al. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J. Clin. Invest. 115, 1636–1643 (2005).
Lanemo Myhrinder, A. et al. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood 111, 3838–3848 (2008).
Ghia, E. M. et al. Use of IGHV3-21 in chronic lymphocytic leukemia is associated with high-risk disease and reflects antigen-driven, post-germinal center leukemogenic selection. Blood 111, 5101–5108 (2008).
Tobin, G. et al. Somatically mutated Ig VH3-21 genes characterize a new subset of chronic lymphocytic leukemia. Blood 99, 2262–2264 (2002).
Potter, K. N. et al. Features of the overexpressed V1-69 genes in the unmutated subset of chronic lymphocytic leukemia are distinct from those in the healthy elderly repertoire. Blood 101, 3082–3084 (2003).
Widhopf, G. F. & Kipps, T. J. Normal B cells express 51p1-encoded Ig heavy chains that are distinct from those expressed by chronic lymphocytic leukemia B cells. J. Immunol. 166, 95–102 (2001).
Catovsky, D. et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. Lancet 370, 230–239 (2007).
Grever, M. R. et al. Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US Intergroup Phase III Trial E2997. J. Clin. Oncol. 25, 799–804 (2007).
Catera, R. et al. Chronic lymphocytic leukemia cells recognize conserved epitopes associated with apoptosis and oxidation. Mol. Med. 14, 665–674 (2008).
Chu, C. C. et al. Chronic lymphocytic leukemia antibodies with a common stereotypic rearrangement recognize nonmuscle myosin heavy chain IIA. Blood 112, 5122–5129 (2008).
Förster, I., Gu, H. & Rajewsky, K. Germline antibody V regions as determinants of clonal persistence and malignant growth in the B cell compartment. EMBO J. 7, 3693–3703 (1988).
Montecino-Rodriguez, E. & Dorshkind, K. New perspectives in B-1 B cell development and function. Trends Immunol. 27, 428–433 (2006).
Fischer, M., Klein, U. & Küppers, R. Molecular single-cell analysis reveals that CD5-positive peripheral blood B cells in healthy humans are characterized by rearranged Vκ genes lacking somatic mutation. J. Clin. Invest. 100, 1667–1676 (1997).
Klein, U., Rajewsky, K. & Küppers, R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 188, 1679–1689 (1998).
Weill, J. C., Weller, S. & Reynaud, C. A. Human marginal zone B cells. Annu. Rev. Immunol. 27, 267–285 (2009).
Willenbrock, K., Jungnickel, B., Hansmann, M. L. & Küppers, R. Human splenic marginal zone B cells lack expression of activation-induced cytidine deaminase. Eur. J. Immunol. 35, 3002–3007 (2005).
Capello, D. et al. Identification of three subgroups of B cell chronic lymphocytic leukemia based upon mutations of BCL-6 and IgV genes. Leukemia 14, 811–815 (2000).
Pasqualucci, L., Neri, A., Baldini, L., Dalla-Favera, R. & Migliazza, A. BCL-6 mutations are associated with immunoglobulin variable heavy chain mutations in B-cell chronic lymphocytic leukemia. Cancer Res. 60, 5644–5648 (2000).
Pasqualucci, L. et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc. Natl Acad. Sci. USA 95, 11816–11821 (1998).
Fukita, Y., Jacobs, H. & Rajewsky, K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity 9, 105–114 (1998).
Allman, D. et al. BCL-6 expression during B-cell activation. Blood 87, 5257–5268 (1996).
Hashimoto, S. et al. Somatic diversification and selection of immunoglobulin heavy and light chain variable region genes in IgG+ CD5+ chronic lymphocytic leukemia B cells. J. Exp. Med. 181, 1507–1517 (1995).
Wakai, M. et al. IgG+, CD5+ human chronic lymphocytic leukemia B cells. Production of IgG antibodies that exhibit diminished autoreactivity and IgG subclass skewing. Autoimmunity 19, 39–48 (1994).
Seifert M, Küppers R. Molecular footprints of a germinal center derivation of human IgM+(IgD+)CD27+ B cells and the dynamics of memory B cell generation. J. Exp. Med. 16 November 2009 (doi:10.1084/jem.20091087)
Sims, G. P. et al. Identification and characterization of circulating human transitional B cells. Blood 105, 4390–4398 (2005).
Matejuk, A. et al. Exclusion of natural autoantibody-producing B cells from IgG memory B cell compartment during T cell-dependent immune responses. J. Immunol. 182, 7634–7643 (2009).
Stevenson, F. K. & Caligaris-Cappio, F. Chronic lymphocytic leukemia: revelations from the B-cell receptor. Blood 103, 4389–4395 (2004).
Rawstron, A. C. et al. Monoclonal B lymphocytes with the characteristics of “indolent” chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood 100, 635–639 (2002).
Nieto, W. G. et al. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood 114, 33–37 (2009).
Rawstron, A. C. et al. Inherited predisposition to CLL is detectable as subclinical monoclonal B-lymphocyte expansion. Blood 100, 2289–2290 (2002).
Landgren, O. et al. B-cell clones as early markers for chronic lymphocytic leukemia. N. Engl. J. Med. 360, 659–667 (2009).
Rawstron, A. C. et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N. Engl. J. Med. 359, 575–583 (2008). The first detailed study on the incidence, genetic profile and clinical course of MBL.
Dagklis, A. et al. The immunoglobulin gene repertoire of low-count CLL-like MBL is different from CLL: diagnostic implications for clinical monitoring. Blood 114, 26–32 (2009).
Marti, G. et al. Overview of monoclonal B-cell lymphocytosis. Br. J. Haematol. 139, 701–708 (2007).
Mertens, D. et al. Allelic silencing at the tumor-suppressor locus 13q14.3 suggests an epigenetic tumor-suppressor mechanism. Proc. Natl Acad. Sci. USA 103, 7741–7746 (2006).
Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 (2001).
Bonci, D. et al. The miR-15a–miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nature Med. 14, 1271–1277 (2008).
Döhner, H. et al. 11q deletions identify a new subset of B-cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood 89, 2516–2522 (1997).
Lavin, M. F. Ataxia–telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nature Rev. Mol. Cell Biol. 9, 759–769 (2008).
Bredemeyer, A. L. et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 442, 466–470 (2006).
Kienle, D. L. et al. Evidence for distinct pathomechanisms in genetic subgroups of chronic lymphocytic leukemia revealed by quantitative expression analysis of cell cycle, activation, and apoptosis-associated genes. J. Clin. Oncol. 23, 3780–3792 (2005).
Kalla, C. et al. Analysis of 11q22–q23 deletion target genes in B-cell chronic lymphocytic leukaemia: evidence for a pathogenic role of NPAT, CUL5, and PPP2R1B. Eur. J. Cancer 43, 1328–1335 (2007).
Stilgenbauer, S. et al. Biologic and clinical markers for outcome after fludarabine (F) or F plus cyclophosphamide (FC) — comprehensive analysis of the CLL4 trial of the GCLLSG. Blood (ASH Annual Meeting Abstracts) 112, 2089 (2008).
Mayr, C. et al. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood 107, 742–751 (2006).
Haferlach, C., Dicker, F., Schnittger, S., Kern, W. & Haferlach, T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgVH status and immunophenotyping. Leukemia 21, 2442–2451 (2007).
Küppers, R. & Dalla-Favera, R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 20, 5580–5594 (2001).
Klein, U. & Dalla-Favera, R. New insights into the phenotype and cell derivation of B cell chronic lymphocytic leukemia. Curr. Top. Microbiol. Immunol. 294, 31–49 (2005).
Di Bernardo, M. C. et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nature Genet. 40, 1204–1210 (2008). These data provide evidence for common, low-penetrance susceptibility loci for CLL.
Lu, R. Interferon regulatory factor 4 and 8 in B-cell development. Trends Immunol. 29, 487–492 (2008).
Rush, L. J. et al. Epigenetic profiling in chronic lymphocytic leukemia reveals novel methylation targets. Cancer Res. 64, 2424–2433 (2004).
Corcoran, M. et al. ZAP-70 methylation status is associated with ZAP-70 expression status in chronic lymphocytic leukemia. Haematologica 90, 1078–1088 (2005).
Bialik, S. & Kimchi, A. The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem. 75, 189–210 (2006).
Byrd, J. C. et al. Depsipeptide (FR901228): a novel therapeutic agent with selective, in vitro activity against human B-cell chronic lymphocytic leukemia cells. Blood 94, 1401–1408 (1999).
Caligaris-Cappio, F. & Ghia, P. Novel insights in chronic lymphocytic leukemia: are we getting closer to understanding the pathogenesis of the disease? J. Clin. Oncol. 26, 4497–4503 (2008).
Messmer, B. T. et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J. Clin. Invest. 115, 755–764 (2005). This elegant study demonstrated that CLL cells had definable and often substantial birth rates. CLL is not a static disease but a dynamic process.
Lagneaux, L., Delforge, A., Bron, D., De Bruyn, C. & Stryckmans, P. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood 91, 2387–2396 (1998).
Ghia, P. et al. Chronic lymphocytic leukemia B cells are endowed with the capacity to attract CD4+, CD40L+ T cells by producing CCL22. Eur. J. Immunol. 32, 1403–1413 (2002).
Burger, J. A. et al. High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood 113, 3050–3058 (2009).
Granziero, L. et al. Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia. Blood 97, 2777–2783 (2001).
Ghia, P. et al. Differential effects on CLL cell survival exerted by different microenvironmental elements. Curr. Top. Microbiol. Immunol. 294, 135–145 (2005).
Hewamana, S. et al. Rel a is an independent biomarker of clinical outcome in chronic lymphocytic leukemia. J. Clin. Oncol. 27, 763–769 (2009).
Ramsay, A. G. et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J. Clin. Invest. 118, 2427–2437 (2008). The demonstration of impaired immunological synapse formation and immune dysfunction in T cells from patients with CLL that can be reversed by immune-modulating drugs.
Patten, P. E. et al. CD38 expression in chronic lymphocytic leukemia is regulated by the tumor microenvironment. Blood 111, 5173–5181 (2008).
Damle, R. N. et al. CD38 expression labels an activated subset within chronic lymphocytic leukemia clones enriched in proliferating B cells. Blood 110, 3352–3359 (2007).
Deaglio, S. et al. CD38 and CD100 lead a network of surface receptors relaying positive signals for B-CLL growth and survival. Blood 105, 3042–3050 (2005).
Burger, J. A. et al. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood 96, 2655–2663 (2000).
Kern, C. et al. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood 103, 679–688 (2004).
Novak, A. J., Bram, R. J., Kay, N. E. & Jelinek, D. F. Aberrant expression of B-lymphocyte stimulator by B chronic lymphocytic leukemia cells: a mechanism for survival. Blood 100, 2973–2979 (2002).
Tsukada, N., Burger, J. A., Zvaifler, N. J. & Kipps, T. J. Distinctive features of “nurselike” cells that differentiate in the context of chronic lymphocytic leukemia. Blood 99, 1030–1037 (2002).
Planelles, L. et al. APRIL promotes B-1 cell-associated neoplasm. Cancer Cell 6, 399–408 (2004).
Chanan-Khan, A. et al. Results of a Phase 1 clinical trial of thalidomide in combination with fludarabine as initial therapy for patients with treatment-requiring chronic lymphocytic leukemia (CLL). Blood 106, 3348–3352 (2005).
Ferrajoli, A. et al. Lenalidomide induces complete and partial remissions in patients with relapsed and refractory chronic lymphocytic leukemia. Blood 111, 5291–5297 (2008).
Dighiero, G. et al. Chlorambucil in indolent chronic lymphocytic leukemia. French Cooperative Group on Chronic Lymphocytic Leukemia. N. Engl. J. Med. 338, 1506–1514 (1998).
Keating, M. J. et al. Early results of a chemoimmunotherapy regimen of fludarabine, cyclophosphamide, and rituximab as initial therapy for chronic lymphocytic leukemia. J. Clin. Oncol. 23, 4079–4088 (2005).
Abrisqueta, P. et al. Improving survival in patients with chronic lymphocytic leukemia (1980–2008): the Hospital Clinic of Barcelona experience. Blood 114, 2044–2050 (2009).
Eichhorst, B. F. et al. Fludarabine plus cyclophosphamide versus fludarabine alone in first-line therapy of younger patients with chronic lymphocytic leukemia. Blood 107, 885–891 (2006).
Wierda, W. G. et al. Characteristics associated with important clinical end points in patients with chronic lymphocytic leukemia at initial treatment. J. Clin. Oncol. 27, 1637–1643 (2009).
Crespo, M. et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N. Engl. J. Med. 348, 1764–1775 (2003). The first demonstration that ZAP70, as detected by flow-cytometric analysis, correlates with IGHV mutational status, disease progression and survival in CLL.
Dreger, P. et al. Indications for allogeneic stem cell transplantation in chronic lymphocytic leukemia: the EBMT transplant consensus. Leukemia 21, 12–17 (2007).
Zenz, T. et al. How little is too much? p53 inactivation: from laboratory cutoff to biological basis of chemotherapy resistance. Leukemia 22, 2257–2258 (2008).
el Rouby, S. et al. p53 gene mutation in B-cell chronic lymphocytic leukemia is associated with drug resistance and is independent of MDR1/MDR3 gene expression. Blood 82, 3452–3459 (1993).
Rossi, D. et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of del17p13: implications for overall survival and chemorefractoriness. Clin. Cancer Res. 15, 995–1004 (2009).
Zenz, T. et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood 113, 3801–3808 (2009).
Pettitt, A. R. et al. p53 dysfunction in B-cell chronic lymphocytic leukemia: inactivation of ATM as an alternative to TP53 mutation. Blood 98, 814–822 (2001).
Stankovic, T. et al. Inactivation of ataxia telangiectasia mutated gene in B-cell chronic lymphocytic leukaemia. Lancet 353, 26–29 (1999).
Stilgenbauer, S. et al. Genomic aberrations, VH mutation status and outcome after fludarabine and cyclophosphamide (FC) or FC plus Rituximab (FCR) in the CLL8 trial. Blood (ASH Annual Meeting Abstracts) 112, 781 (2008).
Callen, E., Nussenzweig, M. C. & Nussenzweig, A. Breaking down cell cycle checkpoints and DNA repair during antigen receptor gene assembly. Oncogene 26, 7759–7764 (2007).
Kojima, K. et al. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood 108, 993–1000 (2006).
Vogler, M. et al. Concurrent up-regulation of BCL-XL and BCL2A1 induces approximately 1000-fold resistance to ABT-737 in chronic lymphocytic leukemia. Blood 113, 4403–4413 (2009).
Buchner, M. et al. Spleen tyrosine kinase is overexpressed and represents a potential therapeutic target in chronic lymphocytic leukemia. Cancer Res. 69, 5424–5432 (2009).
Matutes, E. et al. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia 8, 1640–1645 (1994).
Rassenti, L. Z. et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N. Engl. J. Med. 351, 893–901 (2004).
Kröber, A. et al. Additional genetic high-risk features such as 11q deletion, 17p deletion, and V3-21 usage characterize discordance of ZAP-70 and VH mutation status in chronic lymphocytic leukemia. J. Clin. Oncol. 24, 969–975 (2006).
Montserrat, E. et al. How I treat refractory CLL. Blood 107, 1276–1283 (2006).
Allen, C. D., Okada, T., Tang, H. L. & Cyster, J. G. Imaging of germinal center selection events during affinity maturation. Science 315, 528–531 (2007).
Calin, G. A. et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl Acad. Sci. USA 105, 5166–5171 (2008).
Pedersen, I. M. et al. Protection of CLL B cells by a follicular dendritic cell line is dependent on induction of Mcl-1. Blood 100, 1795–1801 (2002).
Wiestner, A. et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood 101, 4944–4951 (2003).
Roos, G. et al. Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood 111, 2246–2252 (2008).
Shanafelt, T. D. et al. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J. Clin. Oncol. 24, 4634–4641 (2006).
Fabris, S. et al. Molecular and transcriptional characterization of 17p loss in B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer 47, 781–793 (2008).
Zenz, T. et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53–p21 dysfunction, and miR34a in a prospective clinical trial. Blood 114, 2589–2597 (2009).
Zenz, T., Dohner, H. & Stilgenbauer, S. Genetics and risk-stratified approach to therapy in chronic lymphocytic leukemia. Best Pract. Res. Clin. Haematol. 20, 439–453 (2007).
Stilgenbauer, S. et al. Subcutaneous alemtuzumab in fludarabine-refractory chronic lymphocytic leukemia: clinical results and prognostic marker analyses from the CLL2H study of the German Chronic Lymphocytic Leukemia Study Group. J. Clin. Oncol. 27, 3994–4001 (2009).
Acknowledgements
We acknowledge the important contributions of the numerous researchers whose work could not be cited due to space restrictions. The authors are supported in part by the German José Carreras Leukemia Foundation (R06/28v, R06/13 and R08/26f), Else Kröner-Fresenius-Stiftung (P20/07//A11/07), Deutsche Krebshilfe (108,355, 106,142 and 107,239), SBCancer Helmholtz Alliance on Systems Biology and the Global CLL Research Foundation. We thank Antonio Sarno and John Byrd for critical reading of the manuscript. We thank the GCLLSG and the chairman Michael Hallek for long-standing cooperation and support. We thank the European Research Initiative on CLL (ERIC) for ongoing discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors receive research funding and other remuneration from Roche, GSK, Bayer Schering pharma and Celgene. They receive other remuneration from Trubion pharmaceuticals.
Related links
Related links
DATABASES
National Cancer Institute Drug Dictionary
OMIM
FURTHER INFORMATION
Daniel Mertens' laboratory homepage
CLL Global Research Foundation
Department of Internal Medicine III at the University of Ulm
Glossary
- Stereotyped B cell receptors
-
Strikingly similar B cell receptors, which often arise from the use of common H and L chain V region gene segments that share CDR3 structural features (such as their length, amino acid composition and unique amino acid residues at recombination junctions).
- Antigenic drive
-
CLL cells seem to be selected by a limited set of antigenic epitopes at some point in their development. CLL cells are stimulated by the binding of these antigens to the BCR.
- Somatic hypermutation
-
A process that modifies the immunoglobulin variable region genes by introducing mutations into them at a high rate.
- Anergic
-
A state in which B or T cells are unresponsive and cannot be activated by antigen.
- IGHV1-69
-
A specific IGHV gene found at a high frequency in CLLs with unmutated IGHV.
- CpG island
-
A region of DNA with a high density of cytosine phosphoguanine dinucleotides, which are near the transcriptional start sites of 40% of all mammalian genes. Cytosine methylation in CpG islands is generally associated with stable silencing of the associated gene.
- Immunological synapse
-
The supramolecular structure that is established between a T cell and an antigen-presenting cell or B cell.
- Binet stage
-
A clinical staging system most commonly used in Europe based on lymphadenopathy, spleen and liver size and blood count (red cells and platelets).
- Rai classification
-
A clinical staging system most commonly used in the United States.
Rights and permissions
About this article
Cite this article
Zenz, T., Mertens, D., Küppers, R. et al. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 10, 37–50 (2010). https://doi.org/10.1038/nrc2764
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc2764
This article is cited by
-
WNT3 and LEF1 as markers for diagnosis and survival prediction in chronic lymphocytic leukemia patients
memo - Magazine of European Medical Oncology (2023)
-
Sex-specific DNA methylation: impact on human health and development
Molecular Genetics and Genomics (2022)
-
DDX3X: structure, physiologic functions and cancer
Molecular Cancer (2021)
-
Cooperative miRNA-dependent PTEN regulation drives resistance to BTK inhibition in B-cell lymphoid malignancies
Cell Death & Disease (2021)
-
Multi-omics reveals clinically relevant proliferative drive associated with mTOR-MYC-OXPHOS activity in chronic lymphocytic leukemia
Nature Cancer (2021)