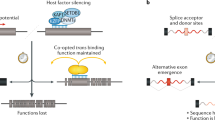
Key Points
-
Much of the human genome is repetitive sequence derived from transposable elements. These include copy-and-paste retrotransposons and cut-and-paste DNA transposons. Only retrotransposons are active as undomesticated mobile DNAs in humans.
-
Ongoing retrotransposition in humans is attributed to long interspersed element-1 (LINE-1; also known as L1). Its activity creates genomic structural variants in human populations and alterations in cancer genomes. Endogenous retroviruses persist as promoter and protein-coding sequences.
-
LINE-1 encodes open reading frame 1p (ORF1p) and ORF2p proteins. ORF1p is an RNA binding protein widely expressed in human malignancies. ORF2p encodes endonuclease and reverse transcriptase activities essential for retrotransposition.
-
LINE-1 activity generates new copies of itself, and also other sequences, including Alu and short interspersed element (SINE)–variable number tandem repeat (VNTR)–Alu (SVA) retrotransposons, pseudogene copies of messenger transcripts and U6 ribosomal RNAs (rRNAs).
-
In cancer, source LINE-1 elements escape genomic DNA methylation and transcriptional repression to contribute new LINE-1 insertions. Different source elements may be active over different phases of the evolution of a cancer.
-
Somatically acquired LINE-1 insertions can cause driver mutations, particularly in gastrointestinal tract tumours, which support high levels of retrotransposition. Distinguishing contributing mutations from inert passenger mutations is an important challenge for the field.
Abstract
Transposable elements give rise to interspersed repeats, sequences that comprise most of our genomes. These mobile DNAs have been historically underappreciated — both because they have been presumed to be unimportant, and because their high copy number and variability pose unique technical challenges. Neither impediment now seems steadfast. Interest in the human mobilome has never been greater, and methods enabling its study are maturing at a fast pace. This Review describes the activity of transposable elements in human cancers, particularly long interspersed element-1 (LINE-1). LINE-1 sequences are self-propagating, protein-coding retrotransposons, and their activity results in somatically acquired insertions in cancer genomes. Altered expression of transposable elements and animation of genomic LINE-1 sequences appear to be hallmarks of cancer, and can be responsible for driving mutations in tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Jurka, J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16, 418–420 (2000).
Boissinot, S., Chevret, P. & Furano, A. V. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17, 915–928 (2000).
Huang, C. R., Burns, K. H. & Boeke, J. D. Active transposition in genomes. Annu. Rev. Genet. 46, 651–675 (2012).
Kazazian, H. H. Jr. et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–166 (1988).
Hancks, D. C. & Kazazian, H. H. Jr. Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9 (2016).
Moran, J. V. et al. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927 (1996).
Ostertag, E. M., Prak, E. T., DeBerardinis, R. J., Moran, J. V. & Kazazian, H. H. Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423 (2000).
Kopera, H. C. et al. LINE-1 cultured cell retrotransposition assay. Methods Mol. Biol. 1400, 139–156 (2016).
Ostertag, E. M. et al. A mouse model of human L1 retrotransposition. Nat. Genet. 32, 655–660 (2002).
An, W. et al. Active retrotransposition by a synthetic L1 element in mice. Proc. Natl Acad. Sci. USA 103, 18662–18667 (2006).
Batzer, M. A. & Deininger, P. L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3, 370–379 (2002).
Xing, J. et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 19, 1516–1526 (2009).
Ewing, A. D. & Kazazian, H. H. Jr. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 20, 1262–1270 (2010).
Ewing, A. D. & Kazazian, H. H. Jr. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 21, 985–990 (2011).
Hormozdiari, F. et al. Alu repeat discovery and characterization within human genomes. Genome Res. 21, 840–849 (2011).
Stewart, C. et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 7, e1002236 (2011).
Sudmant, P. H. et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015).
Richardson, S. R. et al. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol. Spectr. 3, MDNA3-0061–2014 (2015).
Boeke, J. D., Garfinkel, D. J., Styles, C. A. & Fink, G. R. Ty elements transpose through an RNA intermediate. Cell 40, 491–500 (1985).
Dewannieux, M., Esnault, C. & Heidmann, T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48 (2003).
Hancks, D. C., Goodier, J. L., Mandal, P. K., Cheung, L. E. & Kazazian, H. H. Jr. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 20, 3386–3400 (2011).
Raiz, J. et al. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40, 1666–1683 (2012).
Esnault, C., Maestre, J. & Heidmann, T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367 (2000).
Wei, W. et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439 (2001).
Buzdin, A. et al. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of L1. Genomics 80, 402–406 (2002).
Gilbert, N., Lutz, S., Morrish, T. A. & Moran, J. V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25, 7780–7795 (2005).
Garcia-Perez, J. L., Doucet, A. J., Bucheton, A., Moran, J. V. & Gilbert, N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 17, 602–611 (2007).
Doucet, A. J., Droc, G., Siol, O., Audoux, J. & Gilbert, N. U6 snRNA pseudogenes: markers of retrotransposition dynamics in mammals. Mol. Biol. Evol. 32, 1815–1832 (2015).
Schueler, M. G. & Sullivan, B. A. Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genom. Hum. Genet. 7, 301–313 (2006).
Mefford, H. C. & Trask, B. J. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 3, 91–102 (2002).
de Koning, A. P., Gu, W., Castoe, T. A., Batzer, M. A. & Pollock, D. D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7, e1002384 (2011).
Wicker, T. et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (2007).
Giordano, J. et al. Evolutionary history of mammalian transposons determined by genome-wide defragmentation. PLoS Comput. Biol. 3, e137 (2007).
Boissinot, S., Entezam, A., Young, L., Munson, P. J. & Furano, A. V. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 14, 1221–1231 (2004).
Boissinot, S. & Furano, A. V. The recent evolution of human L1 retrotransposons. Cytogenet. Genome Res. 110, 402–406 (2005).
Beck, C. R. et al. LINE-1 retrotransposition activity in human genomes. Cell 141, 1159–1170 (2010). Description of full-length LINE-1 polymorphisms in human populations, and identification of a subset of highly active elements for retrotransposition.
Huang, C. R. et al. Mobile interspersed repeats are major structural variants in the human genome. Cell 141, 1171–1182 (2010).
Iskow, R. C. et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141, 1253–1261 (2010). Demonstration that massively parallel sequencing can be targeted to recover germline and somatically acquired retrotransposon insertions.
Deininger, P. Alu elements: know the SINEs. Genome Biol. 12, 236 (2011).
Witherspoon, D. J. et al. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics 11, 410 (2010).
Witherspoon, D. J. et al. Mobile element scanning (ME-Scan) identifies thousands of novel Alu insertions in diverse human populations. Genome Res. 23, 1170–1181 (2013).
Jurka, J., Zietkiewicz, E. & Labuda, D. Ubiquitous mammalian-wide interspersed repeats (MIRs) are molecular fossils from the mesozoic era. Nucleic Acids Res. 23, 170–175 (1995).
Smit, A. F. & Riggs, A. D. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 23, 98–102 (1995).
Hancks, D. C., Mandal, P. K., Cheung, L. E. & Kazazian, H. H. Jr. The minimal active human SVA retrotransposon requires only the 5′-hexamer and Alu-like domains. Mol. Cell. Biol. 32, 4718–4726 (2012).
Hancks, D. C. & Kazazian, H. H. Jr. SVA retrotransposons: evolution and genetic instability. Semin. Cancer Biol. 20, 234–245 (2010).
Smit, A. F. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 21, 1863–1872 (1993).
Dewannieux, M. et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16, 1548–1556 (2006).
Belshaw, R. et al. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 79, 12507–12514 (2005).
Hughes, J. F. & Coffin, J. M. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl Acad. Sci. USA 101, 1668–1672 (2004).
Turner, G. et al. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11, 1531–1535 (2001).
Smit, A. F. & Riggs, A. D. Tiggers and DNA transposon fossils in the human genome. Proc. Natl Acad. Sci. USA 93, 1443–1448 (1996).
Roth, D. B. & Craig, N. L. VDJ recombination: a transposase goes to work. Cell 94, 411–414 (1998).
Agrawal, A., Eastman, Q. M. & Schatz, D. G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–751 (1998).
Hencken, C. G., Li, X. & Craig, N. L. Functional characterization of an active Rag-like transposase. Nat. Struct. Mol. Biol. 19, 834–836 (2012).
Hiom, K., Melek, M. & Gellert, M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94, 463–470 (1998).
Kapitonov, V. V. & Jurka, J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 3, e181 (2005).
Huang, S. et al. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell 166, 102–114 (2016).
Liu, D. et al. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell. Biol. 27, 1125–1132 (2007).
Majumdar, S., Singh, A. & Rio, D. C. The human THAP9 gene encodes an active P-element DNA transposase. Science 339, 446–448 (2013).
Henssen, A. G. et al. Genomic DNA transposition induced by human PGBD5. Elife 4, e10565 (2015).
Yoder, J. A., Walsh, C. P. & Bestor, T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997).
Siomi, M. C., Sato, K., Pezic, D. & Aravin, A. A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258 (2011).
Imbeault, M., Helleboid, P. Y. & Trono, D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554 (2017).
Jacobs, F. M. et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245 (2014).
Quenneville, S. et al. The KRAB−ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Rep. 2, 766–773 (2012).
Rowe, H. M. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 (2010).
Turelli, P. et al. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 24, 1260–1270 (2014).
Alves, G., Tatro, A. & Fanning, T. Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene 176, 39–44 (1996).
Shukla, R. et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153, 101–111 (2013). Mapping of LINE-1 insertions in hepatocellular carcinoma and tracking of inherited and acquired LINE-1 in non-coding DNA that relate to pathogenesis.
Tubio, J. M. et al. Mobile DNA in cancer: extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345, 1251343 (2014). Demonstration of a set of source elements responsible for LINE-1 retrotransposition with 3′ transduction in human cancers.
Scott, E. C. et al. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 26, 745–755 (2016). Description of a second somatically acquired LINE-1 insertion in the APC gene, which could be traced to a source element that escaped silencing in the normal colon.
Daskalos, A. et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int. J. Cancer 124, 81–87 (2009).
Saito, K. et al. Long interspersed nuclear element 1 hypomethylation is a marker of poor prognosis in stage IA non-small cell lung cancer. Clin. Cancer Res. 16, 2418–2426 (2010).
Ogino, S. et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J. Natl Cancer Inst. 100, 1734–1738 (2008).
Iwagami, S. et al. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann. Surg. 257, 449–455 (2013).
van Hoesel, A. Q. et al. Hypomethylation of LINE-1 in primary tumor has poor prognosis in young breast cancer patients: a retrospective cohort study. Breast Cancer Res. Treat. 134, 1103–1114 (2012).
Harada, K. et al. LINE-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Ann. Surg. Oncol. 22, 1280–1287 (2015).
Pattamadilok, J. et al. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int. J. Gynecol. Cancer 18, 711–717 (2008).
Hall, L. L. et al. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell 156, 907–919 (2014).
Skowronski, J., Fanning, T. G. & Singer, M. F. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell. Biol. 8, 1385–1397 (1988).
Belancio, V. P., Hedges, D. J. & Deininger, P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 34, 1512–1521 (2006).
Belancio, V. P., Roy-Engel, A. M. & Deininger, P. The impact of multiple splice sites in human L1 elements. Gene 411, 38–45 (2008).
Deininger, P. & Belancio, V. P. Detection of LINE-1 RNAs by northern blot. Methods Mol. Biol. 1400, 223–236 (2016).
Philippe, C. et al. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife 5, e13926 (2016). Description of a tailored approach to RNA-seq and ChIP-sequencing, which can be used to identify loci expressing LINE-1 unit transcripts.
Deininger, P. et al. A comprehensive approach to expression of L1 loci. Nucleic Acids Res. 45, e31 (2016).
Pickeral, O. K., Makalowski, W., Boguski, M. S. & Boeke, J. D. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10, 411–415 (2000).
Goodier, J. L., Ostertag, E. M. & Kazazian, H. H. Jr. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 9, 653–657 (2000).
Matlik, K., Redik, K. & Speek, M. L1 antisense promoter drives tissue-specific transcription of human genes. J. Biomed. Biotechnol. 2006, 71753 (2006).
Nigumann, P., Redik, K., Matlik, K. & Speek, M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79, 628–634 (2002).
Criscione, S. W. et al. Genome-wide characterization of human L1 antisense promoter-driven transcripts. BMC Genomics 17, 463 (2016).
Roman-Gomez, J. et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene 24, 7213–7223 (2005).
Lin, L. et al. Multiple forms of genetic instability within a 2-Mb chromosomal segment of 3q26.3−q27 are associated with development of esophageal adenocarcinoma. Genes Chromosomes Cancer 45, 319–331 (2006).
Cruickshanks, H. A. & Tufarelli, C. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 94, 397–406 (2009).
Weber, B., Kimhi, S., Howard, G., Eden, A. & Lyko, F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene 29, 5775–5784 (2010).
Wolff, E. M. et al. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 6, e1000917 (2010).
Hur, K. et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 63, 635–646 (2014).
Denli, A. M. et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163, 583–593 (2015).
Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 9, 397–405 (2008).
Lamprecht, B. et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 16, 571–579 (2010).
Babaian, A. et al. Onco-exaptation of an endogenous retroviral LTR drives IRF5 expression in Hodgkin lymphoma. Oncogene 35, 2542–2546 (2016).
Scarfo, I. et al. Identification of a new subclass of ALK-negative ALCL expressing aberrant levels of ERBB4 transcripts. Blood 127, 221–232 (2016).
Lock, F. E. et al. Distinct isoform of FABP7 revealed by screening for retroelement-activated genes in diffuse large B-cell lymphoma. Proc. Natl Acad. Sci. USA 111, E3534–E3543 (2014).
Babaian, A. & Mager, D. L. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 7, 24 (2016).
Cruickshanks, H. A. et al. Expression of a large LINE-1-driven antisense RNA is linked to epigenetic silencing of the metastasis suppressor gene TFPI-2 in cancer. Nucleic Acids Res. 41, 6857–6869 (2013).
Domansky, A. N. et al. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472, 191–195 (2000).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Holmes, S. E., Singer, M. F. & Swergold, G. D. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267, 19765–19768 (1992).
Dombroski, B. A., Mathias, S. L., Nanthakumar, E., Scott, A. F. & Kazazian, H. H. Jr. Isolation of an active human transposable element. Science 254, 1805–1808 (1991).
Luan, D. D., Korman, M. H., Jakubczak, J. L. & Eickbush, T. H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72, 595–605 (1993).
Christensen, S. M., Bibillo, A. & Eickbush, T. H. Role of the Bombyx mori R2 element N-terminal domain in the target-primed reverse transcription (TPRT) reaction. Nucleic Acids Res. 33, 6461–6468 (2005).
Feng, Q., Moran, J. V., Kazazian, H. H. Jr & Boeke, J. D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916 (1996).
Mathias, S. L. et al. Reverse transcriptase encoded by a human transposable element. Science 254, 1808–1810 (1991).
Brouha, B. et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA 100, 5280–5285 (2003). Study that compared commonly occurring LINE-1 insertions for retrotransposition activity in vitro and established the term hot LINE-1.
Ullu, E. & Tschudi, C. Alu sequences are processed 7SL RNA genes. Nature 312, 171–172 (1984).
Zietkiewicz, E., Richer, C., Sinnett, D. & Labuda, D. Monophyletic origin of Alu elements in primates. J. Mol. Evol. 47, 172–182 (1998).
Ahl, V., Keller, H., Schmidt, S. & Weichenrieder, O. Retrotransposition and crystal structure of an Alu RNP in the ribosome-stalling conformation. Mol. Cell 60, 715–727 (2015).
Doucet, A. J., Wilusz, J. E., Miyoshi, T., Liu, Y. & Moran, J. V. A. 3′ Poly(A) tract is required for LINE-1 retrotransposition. Mol. Cell 60, 728–741 (2015).
Lee, E. et al. Landscape of somatic retrotransposition in human cancers. Science 337, 967–971 (2012). Demonstration of the different types of human cancer that support LINE-1 retrotransposition.
Hohjoh, H. & Singer, M. F. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J. Mol. Biol. 271, 7–12 (1997).
Hohjoh, H. & Singer, M. F. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16, 6034–6043 (1997).
Martin, S. L., Branciforte, D., Keller, D. & Bain, D. L. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl Acad. Sci. USA 100, 13815–13820 (2003).
Khazina, E. et al. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 18, 1006–1014 (2011).
Hohjoh, H. & Singer, M. F. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15, 630–639 (1996).
Kulpa, D. A. & Moran, J. V. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 14, 3237–3248 (2005).
Taylor, M. S. et al. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155, 1034–1048 (2013).
Rodic, N. et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am. J. Pathol. 184, 1280–1286 (2014). A survey of different types of human cancer that aberrantly express LINE-1-encoded ORF1p.
Leibold, D. M. et al. Translation of LINE-1 DNA elements in vitro and in human cells. Proc. Natl Acad. Sci. USA 87, 6990–6994 (1990).
Ardeljan, D., Taylor, M. S., Ting, D. T. & Burns, K. H. The human long interspersed element-1 retrotransposon: an emerging biomarker of neoplasia. Clin. Chem. http://dx.doi.org/10.1373/clinchem.2016.257444 (2017).
Achanta, P. et al. Somatic retrotransposition is infrequent in glioblastomas. Mob. DNA 7, 22 (2016).
Wylie, A. et al. p53 genes function to restrain mobile elements. Genes Dev. 30, 64–77 (2016).
Rodic, N. et al. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat. Med. 21, 1060–1064 (2015). Profile of LINE-1 insertion sites in primary and metastatic PDAC samples, inferring when and where new insertions were acquired.
Goodier, J. L., Zhang, L., Vetter, M. R. & Kazazian, H. H. Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483 (2007).
Sokolowski, M. et al. Development of a monoclonal antibody specific to the endonuclease domain of the human LINE-1 ORF2 protein. Mob. DNA 5, 29 (2014).
De Luca, C. et al. Enhanced expression of LINE-1-encoded ORF2 protein in early stages of colon and prostate transformation. Oncotarget 7, 4048–4061 (2016).
Denne, M. et al. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J. Virol. 81, 5607–5616 (2007).
Mangeney, M., de Parseval, N., Thomas, G. & Heidmann, T. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J. Gen. Virol. 82, 2515–2518 (2001).
Miki, Y. et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 52, 643–645 (1992). Description of the discovery of a somatically acquired LINE-1 insertion in the APC gene that causes colon cancer.
Fodde, R., Smits, R. & Clevers, H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer 1, 55–67 (2001).
Groden, J. et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66, 589–600 (1991).
Joslyn, G. et al. Identification of deletion mutations and three new genes at the familial polyposis locus. Cell 66, 601–613 (1991).
Nishisho, I. et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 253, 665–669 (1991).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Powell, S. M. et al. APC mutations occur early during colorectal tumorigenesis. Nature 359, 235–237 (1992).
Cost, G. J., Feng, Q., Jacquier, A. & Boeke, J. D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21, 5899–5910 (2002).
Jurka, J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl Acad. Sci. USA 94, 1872–1877 (1997).
Ostertag, E. M. & Kazazian, H. H. Jr. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 11, 2059–2065 (2001).
Solyom, S. et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 22, 2328–2338 (2012).
Doucet-O'Hare, T. T. et al. LINE-1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc. Natl Acad. Sci. USA 112, E4894–E4900 (2015).
Doucet-O'Hare, T. T. et al. Somatically acquired LINE-1 insertions in normal esophagus undergo clonal expansion in esophageal squamous cell carcinoma. Hum. Mutat. 37, 942–954 (2016).
Ewing, A. D. et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 25, 1536–1545 (2015).
Helman, E. et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 24, 1053–1063 (2014).
Tang, Z. et al. Human transposon insertion profiling: analysis, visualization and identification of somatic LINE-1 insertions in ovarian cancer. Proc. Natl Acad. Sci. USA 114, E733–E740 (2017).
Carreira, P. E. et al. Evidence for L1-associated DNA rearrangements and negligible L1 retrotransposition in glioblastoma multiforme. Mob. DNA 7, 21 (2016).
Holmes, S. E., Dombroski, B. A., Krebs, C. M., Boehm, C. D. & Kazazian, H. H. Jr. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat. Genet. 7, 143–148 (1994).
Moran, J. V., DeBerardinis, R. J. & Kazazian, H. H. Jr. Exon shuffling by L1 retrotransposition. Science 283, 1530–1534 (1999).
Macfarlane, C. M. et al. Transduction-specific ATLAS reveals a cohort of highly active L1 retrotransposons in human populations. Hum. Mutat. 34, 974–985 (2013).
Ewing, A. D. et al. Retrotransposition of gene transcripts leads to structural variation in mammalian genomes. Genome Biol. 14, R22 (2013).
Cooke, S. L. et al. Processed pseudogenes acquired somatically during cancer development. Nat. Commun. 5, 3644 (2014).
Mir, A. A., Philippe, C. & Cristofari, G. euL1db: the European database of L1HS retrotransposon insertions in humans. Nucleic Acids Res. 43, D43–D47 (2015).
Wheelan, S. J., Aizawa, Y., Han, J. S. & Boeke, J. D. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 15, 1073–1078 (2005).
Sorek, R., Ast, G. & Graur, D. Alu-containing exons are alternatively spliced. Genome Res. 12, 1060–1067 (2002).
Lev-Maor, G. et al. Intronic Alus influence alternative splicing. PLoS Genet. 4, e1000204 (2008).
Symer, D. E. et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110, 327–338 (2002).
Gilbert, N., Lutz-Prigge, S. & Moran, J. V. Genomic deletions created upon LINE-1 retrotransposition. Cell 110, 315–325 (2002).
Lehrman, M. A. et al. Mutation in LDL receptor: Alu−Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science 227, 140–146 (1985).
Rahbari, R. & Badge, R. M. Combining amplification typing of L1 active subfamilies (ATLAS) with high-throughput sequencing. Methods Mol. Biol. 1400, 95–106 (2016).
Streva, V. A. et al. Sequencing, identification and mapping of primed L1 elements (SIMPLE) reveals significant variation in full length L1 elements betweenindividuals. BMC Genomics 16, 220 (2015).
Quentin, Y. Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res. 20, 487–493 (1992).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Johns Hopkins University School of Medicine has licensed long interspersed element-1 (LINE-1) open reading frame 1p (ORF1p) monoclonal antibodies developed in the Burns laboratory for commercial production (K.H.B.).
Related links
FURTHER INFORMATION
Glossary
- Retrotransposition
-
A mechanism used by transposable elements to copy RNA intermediates to genomic DNA.
- Centromeric satellites
-
Arrays of tandem, simple repeats at the regions of chromosomes that attach to the mitotic spindle.
- Human reference genome assembly
-
A version of the human genome, for example, the human Dec. 2013 (GRCh38/hg38) assembly. Structural variants caused by mobile element insertions are not consistently incorporated.
- Source element
-
Full-length genomic long interspersed element-1 (LINE-1) sequence capable of retrotransposition.
- PIWI-interacting RNAs
-
(piRNAs). Small RNAs (26–31 nucleotides) that bind to PIWI family proteins and have key roles in silencing retroelements.
- Short read alignments
-
Alignments of short reads from massively parallel sequencing (MPS) studies to a reference sequence; for example, to the human reference genome assembly.
- Target primed reverse transcription
-
(TPRT). The molecular process executed by long interspersed element-1 (LINE-1) open reading frame 2p (ORF2p), using a strand of target site DNA to prime reverse transcription of an RNA.
- Cis-preference
-
The tendency of long interspersed element-1 (LINE-1)-encoded open reading frame 2p (ORF2p) to associate with and reverse transcribe the RNA strand that encoded the protein. This opposes other RNA species that co-opt ORF2p by association in trans.
Rights and permissions
About this article
Cite this article
Burns, K. Transposable elements in cancer. Nat Rev Cancer 17, 415–424 (2017). https://doi.org/10.1038/nrc.2017.35
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc.2017.35
This article is cited by
-
BORIS/CTCFL epigenetically reprograms clustered CTCF binding sites into alternative transcriptional start sites
Genome Biology (2024)
-
Towards targeting transposable elements for cancer therapy
Nature Reviews Cancer (2024)
-
Jump-starting life: balancing transposable element co-option and genome integrity in the developing mammalian embryo
EMBO Reports (2024)
-
Regulation and function of transposable elements in cancer genomes
Cellular and Molecular Life Sciences (2024)
-
A human-specific insertion promotes cell proliferation and migration by enhancing TBC1D8B expression
Science China Life Sciences (2024)