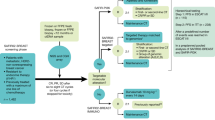
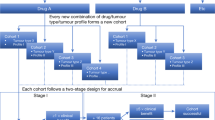
Abstract
Genomic profiling of tumours in patients in clinical trials enables rapid testing of multiple hypotheses to confirm which genomic events determine likely responder groups for targeted agents. A key challenge of this new capability is defining which specific genomic events should be classified as 'actionable' (that is, potentially responsive to a targeted therapy), especially when looking for early indications of patient subgroups likely to be responsive to new drugs. This Opinion article discusses some of the different approaches being taken in early clinical development to define actionable mutations, and describes our strategy to address this challenge in early-stage exploratory clinical trials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Gridelli, C. et al. Non-small-cell lung cancer. Nat. Rev. Dis. Primers 1, 15009 (2015).
Simon, R. & Roychowdhury, S. Implementing personalized cancer genomics in clinical trials. Nat. Rev. Drug Discov. 12, 358–369 (2013).
Herbst, R. S. et al. Lung master protocol (Lung-MAP)—a biomarker-driven protocol for accelerating development of therapies for squamous cell lung cancer: SWOG S1400. Clin. Cancer Res. 21, 1514–1524 (2015).
Sleijfer, S., Bogaerts, J. & Siu, L. L. Designing transformative clinical trials in the cancer genome era. J. Clin. Oncol. 31, 1834–1841 (2013).
Andre, F. et al. Prioritizing targets for precision cancer medicine. Ann. Oncol. 25, 2295–2303 (2014).
Hollingsworth, S. Precision medicine in oncology drug development – a pharma perspective. Drug Discov. Today 20, 1455–1463 (2015).
Van Allen, E. M. et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 (2014).
Chau, N. G. & Lorch, J. H. Exceptional responders inspire change: lessons for drug development from the bedside to the bench and back. Oncologist 20, 699–701 (2015).
Biankin, A. V., Piantadosi, S. & Hollingsworth, S. J. Patient-centric trials for therapeutic development in precision oncology. Nature 526, 361–370 (2015).
Stratton, M. R., Campbell, P. J. & Futreal, P. A. The cancer genome. Nature 458, 719–724 (2009).
Ulahannan, D., Kovac, M. B., Mulholland, P. J., Cazier, J. B. & Tomlinson, I. Technical and implementation issues in using next-generation sequencing of cancers in clinical practice. Br. J. Cancer 109, 827–835 (2013).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Nazarian, R. et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT01888601 (2015).
Thress, K. S. et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat. Med. 21, 560–562 (2015).
Siravegna, G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21, 795–801 (2015).
Dougherty, B. et al. Exploratory analyses suggest ovarian tumors with somatic or germline loss of function mutations in BRCA1 or BRCA2 are biologically similar and sensitive to PARP inhibition. Cancer Res. 75, 611 (abstract 611) (2015).
Jones, S. et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci. Transl Med. 7, 283ra53 (2015).
Rodon, J. et al. Challenges in initiating and conducting personalized cancer therapy trials: perspectives from WINTHER, a Worldwide Innovative Network (WIN) Consortium trial. Ann. Oncol. 26, 1791–1798 (2015).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Mertens, F., Johansson, B., Fioretos, T. & Mitelman, F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer 15, 371–381 (2015).
Kong-Beltran, M. et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 66, 283–289 (2006).
Onozato, R. et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J. Thorac. Oncol. 4, 5–11 (2009).
Ng, K. P. et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat. Med. 18, 521–528 (2012).
Antonarakis, E. S. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014).
Szabo, C., Masiello, A., Ryan, J. F. & Brody, L. C. The breast cancer information core: database design, structure, and scope. Hum. Mutat. 16, 123–131 (2000).
Baralle, D. & Baralle, M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 42, 737–748 (2005).
Forbes, S. A. et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Plon, S. E. et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 29, 1282–1291 (2008).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
International Cancer Genome Consortium. International network of cancer genome projects. Nature 464, 993–998 (2010).
Janne, P. A. et al. Impact of KRAS codon subtypes from a randomised phase II trial of selumetinib plus docetaxel in KRAS mutant advanced non-small-cell lung cancer. Br. J. Cancer 113, 199–203 (2015).
Skoulidis, F. et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 5, 860–877 (2015).
Chibon, F. Cancer gene expression signatures – the rise and fall? Eur. J. Cancer 49, 2000–2009 (2013).
Hollander, M. C., Blumenthal, G. M. & Dennis, P. A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer 11, 289–301 (2011).
Rehm, H. L. et al. ClinGen—the clinical genome resource. N. Engl. J. Med. 372, 2235–2242 (2015).
Martelotto, L. G. et al. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol. 15, 484 (2014).
Miosge, L. A. et al. Comparison of predicted and actual consequences of missense mutations. Proc. Natl Acad. Sci. USA 112, E5189–5198 (2015).
1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Pennington, K. P. et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 20, 764–775 (2014).
Richards, C. S. et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet. Med. 10, 294–300 (2008).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Petitjean, A. et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 28, 622–629 (2007).
Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1031 (2013).
Pritchard, C. C. et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J. Mol. Diagn. 16, 56–67 (2014).
Van Allen, E. M. et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 20, 682–688 (2014).
Meric-Bernstam, F. et al. A decision support framework for genomically informed investigational cancer therapy. J. Natl. Cancer Inst. 107, djv098 (2015).
Hovelson, D. H. et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia 17, 385–399 (2015).
McNeil, C. NCI-MATCH launch highlights new trial design in precision-medicine era. J. Natl. Cancer Inst. 107, djv193 (2015).
NCI-Molecular Analysis for Therapy Choice (NCI-MATCH) trial. NIH National Cancer Institute [online], (2015).
Sukhai, M. A. et al. A classification system for clinical relevance of somatic variants identified in molecular profiling of cancer. Genet. Med. 18, 128–136 (2016).
US Food and Drug Administration. FDA approves Lynparza to treat advanced ovarian cancer. FDA [online], (2014).
Middleton, G. et al. The UK National Lung Matrix Trial: translating the biology of stratification in advanced non-small cell lung cancer. Ann. Oncol. 26, 2464–2469 (2015).
American Association for Cancer Research (AACR). Project GENIE. http://www.aacr.org/genie (2015).
NCI and the precision medicine initiative. NIH National Cancer Institute [online], (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02087176 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02087241 (2015).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01357161 (2015).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer. Discov. 2, 401–404 (2012).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors are employees of AstraZeneca and hold shares in the company.
Related links
Rights and permissions
About this article
Cite this article
Carr, T., McEwen, R., Dougherty, B. et al. Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer 16, 319–329 (2016). https://doi.org/10.1038/nrc.2016.35
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc.2016.35
This article is cited by
-
Candidate mechanisms of acquired resistance to first-line osimertinib in EGFR-mutated advanced non-small cell lung cancer
Nature Communications (2023)
-
Precision neuro-oncology: a pilot analysis of personalized treatment in recurrent glioma
Journal of Cancer Research and Clinical Oncology (2023)
-
Anti-cancer treatment schedule optimization based on tumor dynamics modelling incorporating evolving resistance
Scientific Reports (2022)
-
A chemogenomic approach to identify personalized therapy for patients with relapse or refractory acute myeloid leukemia: results of a prospective feasibility study
Blood Cancer Journal (2020)
-
Bicistronic transfer of CDKN2A and p53 culminates in collaborative killing of human lung cancer cells in vitro and in vivo
Gene Therapy (2020)