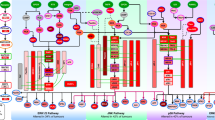
Key Points
-
Application of high-throughput omics technologies has provided important and complementary insights into dysregulation of protein kinases in human cancer and their functional roles.
-
By interrogating a series of recent studies aimed at identifying cancer 'driver' genes causally implicated in cancer development from analysis of cancer genomes, we have identified a list of 91 protein kinases that represent likely cancer drivers. However, the number of protein kinases subject to functionally relevant mutations is likely to be greater, in part reflecting the incidence of non-recurrent mutations that affect signalling networks in a highly context-dependent manner.
-
The list of high-confidence protein kinase cancer drivers includes kinases with well-established roles in cancer development, such as various receptor tyrosine kinases, as well as novel oncogenes and tumour suppressors, such as TATA box binding protein-associated factor 1 (TAF1) and never in mitosis A-related kinase 9 (NEK9).
-
Approximately three-quarters of the high-confidence drivers are either targets for US Food and Drug Administration-approved therapies or amenable to therapeutic targeting through repurposing of existing therapies or further development of those at the preclinical stage. However, there is a substantial need to develop therapeutic strategies that exploit the vulnerabilities conferred by loss of function alterations in specific tumour suppressor kinases.
-
High-throughput analysis of cancer proteomes and sub-proteomes by application of reverse phase protein arrays and mass spectrometry has identified cancer-associated perturbations in protein kinase expression and activity that would not be detected by genomic and transcriptomic approaches. This reflects post-transcriptional regulation of kinase activity at the level of protein expression and post-translational modification.
-
Novel insights provided by proteomics approaches include identification of patterns of kinase activation conserved across various different cancers, the determination of specific tyrosine kinases implicated in non-small-cell lung cancer and basal breast cancer, and characterization of kinome remodelling in response to administration of particular targeted therapies.
-
Functional genomic approaches enable characterization of context-dependent cellular dependency on particular protein kinases and identification of protein kinases that sensitize cancer cells to specific therapies.
-
Integration of particular omics approaches often provides novel insights not provided by individual methodologies, and enables a network-level understanding of kinase signalling that can lead to novel therapeutic opportunities.
Abstract
Over the past decade, rapid advances in genomics, proteomics and functional genomics technologies that enable in-depth interrogation of cancer genomes and proteomes and high-throughput analysis of gene function have enabled characterization of the kinome 'at large' in human cancers, providing crucial insights into how members of the protein kinase superfamily are dysregulated in malignancy, the context-dependent functional role of specific kinases in cancer and how kinome remodelling modulates sensitivity to anticancer drugs. The power of these complementary approaches, and the insights gained from them, form the basis of this Analysis article.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Spector, D. H., Varmus, H. E. & Bishop, J. M. Nucleotide sequences related to the transforming gene of avian sarcoma virus are present in DNA of uninfected vertebrates. Proc. Natl Acad. Sci. USA 75, 4102–4106 (1978).
Hunter, T. Treatment for chronic myelogenous leukemia: the long road to imatinib. J. Clin. Invest. 117, 2036–2043 (2007).
Chapman, P. B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011).
Hudis, C. A. Trastuzumab—mechanism of action and use in clinical practice. N. Engl. J. Med. 357, 39–51 (2007).
Futreal, P. A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004). This article describes the first census of genes causally implicated in cancer. Although more than a decade old and preceding large-scale cancer genome sequencing studies, this article still provides important insights into the type of protein domain that characterizes 'cancer genes'.
Greenman, C. et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007).
Kan, Z. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). This article identifies the crucial issue of heterogeneity of mutation rates within and across cancer types as well as across the genome, and describes the development of an analytical approach, MutSigCV, that addresses this problem and enables the identification of genuine cancer driver genes.
Dees, N. D. et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). This study addresses the power of genomic approaches to generate a comprehensive catalogue of cancer driver genes by analysing mutation data for almost 5,000 cancers across 21 tumour types. In addition to identifying novel cancer drivers, it enabled estimation of the sample size required to generate a complete list of drivers for each type of cancer.
Bolli, N. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 5, 2997 (2014).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
The Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159, 676–690 (2014).
The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014).
The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
Guan, J., Gupta, R. & Filipp, F. V. Cancer systems biology of TCGA SKCM: efficient detection of genomic drivers in melanoma. Sci. Rep. 5, 7857 (2015).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015).
The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). This article has been selected from the many published TCGA studies, as it highlights how large-scale cancer genome sequencing can not only identify novel significantly mutated genes in a given cancer, but also determine the different mutational profiles associated with particular subtypes of that malignancy, in this case the association of MAP3K1 and MAP2K4 mutations with luminal A breast cancer subtype. The article also demonstrates the power of integrated genomic and proteomic analyses.
The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014).
Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Tagliabracci, V. S., Pinna, L. A. & Dixon, J. E. Secreted protein kinases. Trends Biochem. Sci. 38, 121–130 (2013).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Rubio-Perez, C. et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 27, 382–396 (2015).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013). This article describes the application of a computational approach to the prediction of cancer drivers from mutational signatures in tumours. Importantly, it reveals that the distribution of oncogenes and tumour suppressor genes on chromosomes can predict the patterns of aneuploidy and CNA found in cancer genomes.
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Yoshihara, K. et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 34, 4845–4854 (2015).
Owens, P. et al. Disruption of bone morphogenetic protein receptor 2 (BMPR2) in mammary tumors promotes metastases through cell autonomous and paracrine mediators. Proc. Natl Acad. Sci. USA 109, 2814–2819 (2012).
Togashi, Y. et al. Homozygous deletion of the activin A receptor, type IB gene is associated with an aggressive cancer phenotype in pancreatic cancer. Mol. Cancer 13, 126 (2014).
Wang, J. Y. The capable ABL: what is its biological function? Mol. Cell. Biol. 34, 1188–1197 (2014).
Wagner, E. F. & Nebreda, A. R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 9, 537–549 (2009).
Smith, S. C. et al. A gemcitabine sensitivity screen identifies a role for NEK9 in the replication stress response. Nucleic Acids Res. 42, 11517–11527 (2014).
Sonoshita, M. et al. Promotion of colorectal cancer invasion and metastasis through activation of NOTCH-DAB1-ABL-RHOGEF protein TRIO. Cancer Discov. 5, 198–211 (2015).
Nakanishi, Y. et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet. 45, 98–103 (2013).
Blazek, D. et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 25, 2158–2172 (2011).
Ekumi, K. M. et al. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic Acids Res. 43, 2575–2589 (2015).
Bhattacharya, S. et al. Structural and functional insight into TAF1–TAF7, a subcomplex of transcription factor II D. Proc. Natl Acad. Sci. USA 111, 9103–9108 (2014).
Wu, Y. et al. Phosphorylation of p53 by TAF1 inactivates p53-dependent transcription in the DNA damage response. Mol. Cell 53, 63–74 (2014).
Kloet, S. L., Whiting, J. L., Gafken, P., Ranish, J. & Wang, E. H. Phosphorylation-dependent regulation of cyclin D1 and cyclin A gene transcription by TFIID subunits TAF1 and TAF7. Mol. Cell. Biol. 32, 3358–3369 (2012).
Tavassoli, P. et al. TAF1 differentially enhances androgen receptor transcriptional activity via its N-terminal kinase and ubiquitin-activating and -conjugating domains. Mol. Endocrinol. 24, 696–708 (2010).
Antal, C. E. et al. Cancer-associated protein kinase C mutations reveal kinase's role as tumor suppressor. Cell 160, 489–502 (2015).
Creixell, P. et al. Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell 163, 202–217 (2015). This article describes the development of a computational approach termed ReKINect, which is capable of identifying mutations in cancer cells that are predicted to perturb intracellular signalling networks. These include mutations that alter protein kinase substrate selectivity or cause kinase activation or inactivation.
Cagnol, S. & Rivard, N. Oncogenic KRAS and BRAF activation of the MEK/ERK signaling pathway promotes expression of dual-specificity phosphatase 4 (DUSP4/MKP2) resulting in nuclear ERK1/2 inhibition. Oncogene 32, 564–576 (2013).
Akbani, R. et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 5, 3887 (2014). This article describes proteomic characterization of almost 3,500 specimens across 11 cancer types using RPPAs. It highlights how proteomics can determine changes in protein expression and activation not predicted by genomics or transcriptomics, such as HER2 overexpression, and can identify patterns of protein expression and activation shared across different cancer types and that correlate with patient outcome within tumour types.
Al-Ejeh, F. et al. Kinome profiling reveals breast cancer heterogeneity and identifies targeted therapeutic opportunities for triple negative breast cancer. Oncotarget 5, 3145–3158 (2014).
Zhang, B. et al. Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 (2014). This article reports the first application of MS-based proteomics to characterize protein expression in an unbiased fashion in a large cohort of cancer specimens previously subjected to genomic analyses by TCGA. The paper highlights discordance between mRNA and protein levels and demonstrates how proteomics can be used to identify drivers on colorectal cancer amplicons, such as SRC.
Sharma, K. et al. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8, 1583–1594 (2014).
Britton, D. et al. Quantification of pancreatic cancer proteome and phosphorylome: indicates molecular events likely contributing to cancer and activity of drug targets. PLoS ONE 9, e90948 (2014).
Parker, R., Clifton-Bligh, R. & Molloy, M. P. Phosphoproteomics of MAPK inhibition in BRAF-mutated cells and a role for the lethal synergism of dual BRAF and CK2 inhibition. Mol. Cancer Ther. 13, 1894–1906 (2014).
Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007). This study was the first to apply MS-based phosphotyrosine profiling across a large cohort of cancer specimens. It identified novel cancer drivers in NSCLC, including ALK and ROS fusion proteins, and also highlighted different subtypes of this malignancy characterized by distinct tyrosine phosphorylation patterns.
Yoshida, T. et al. Tyrosine phosphoproteomics identifies both codrivers and cotargeting strategies for T790M-related EGFR-TKI resistance in non-small cell lung cancer. Clin. Cancer Res. 20, 4059–4074 (2014).
Bai, Y. et al. Phosphoproteomics identifies driver tyrosine kinases in sarcoma cell lines and tumors. Cancer Res. 72, 2501–2511 (2012).
Hochgrafe, F. et al. Tyrosine phosphorylation profiling reveals the signaling network characteristics of basal breast cancer cells. Cancer Res. 70, 9391–9401 (2010).
Croucher, D. R. et al. Involvement of Lyn and the atypical kinase SgK269/PEAK1 in a basal breast cancer signaling pathway. Cancer Res. 73, 1969–1980 (2013).
Lee, B. Y. et al. Phosphoproteomic profiling identifies focal adhesion kinase as a mediator of docetaxel resistance in castrate-resistant prostate cancer. Mol. Cancer Ther. 13, 190–201 (2014).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Guo, A. et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteom. 13, 372–387 (2014).
Moritz, A. et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci. Signal. 3, ra64 (2010).
Stokes, M. P. et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl Acad. Sci. USA 104, 19855–19860 (2007).
Bantscheff, M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 (2007).
Daub, H. et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 (2008).
Zhang, L. et al. Characterization of the novel broad-spectrum kinase inhibitor CTx-0294885 as an affinity reagent for mass spectrometry-based kinome profiling. J. Proteome Res. 12, 3104–3116 (2013).
Xiao, Y., Guo, L. & Wang, Y. A targeted quantitative proteomics strategy for global kinome profiling of cancer cells and tissues. Mol. Cell. Proteom. 13, 1065–1075 (2014).
Duncan, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012). This article highlights the use of MS-based proteomics to characterize global changes in the expressed kinome in response to drug treatment. It demonstrates that MEK inhibition in TNBC leads to kinome reprogramming that includes increased expression and activation of particular RTKs, leading to resistance to small molecule MEK inhibitors.
Stuhlmiller, T. J. et al. Inhibition of lapatinib-induced kinome reprogramming in ERBB2-positive breast cancer by targeting BET family bromodomains. Cell Rep. 11, 390–404 (2015).
Moghaddas Gholami, A. et al. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 4, 609–620 (2013).
Bollag, G. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599 (2010).
Kopetz, S. et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J. Clin. Oncol. 28 (15_suppl.), 3534 (2010).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012). This article highlights how functional genomics can be used to identify strategies to overcome drug resistance. In this case, a shRNA screen was used to determine that EGFR mediates resistance to vemurafenib in colorectal cancer, leading to clinical trials in which both EGFR and BRAF are targeted in this malignancy.
Grueneberg, D. A. et al. Kinase requirements in human cells: I. Comparing kinase requirements across various cell types. Proc. Natl Acad. Sci. USA 105, 16472–16477 (2008).
Fedorov, O., Muller, S. & Knapp, S. The (un)targeted cancer kinome. Nat. Chem. Biol. 6, 166–169 (2010).
The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Bajrami, I. et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 74, 287–297 (2014).
Yaeger, R. et al. Pilot trial of combined BRAF and EGFR inhibition in BRAF-mutant metastatic colorectal cancer patients. Clin. Cancer Res. 21, 1313–1320 (2015).
Cheung, H. W. et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl Acad. Sci. USA 108, 12372–12377 (2011).
Marcotte, R. et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2, 172–189 (2012).
Brough, R. et al. Functional viability profiles of breast cancer. Cancer Discov. 1, 260–273 (2011). References 77–79 highlight how functional screens across large panels of cancer cell lines can identify genetic dependencies associated with particular cancer types and molecular characteristics. For example, the Brough et al . study identified a dependency on TTK in PTEN-deficient breast cancer cells, identifying a vulnerability that could be exploited for therapeutic development.
Cowley, G. S. et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data 1, 140035 (2014).
Lara, R. et al. An siRNA screen identifies RSK1 as a key modulator of lung cancer metastasis. Oncogene 30, 3513–3521 (2011).
Weissmueller, S. et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell 157, 382–394 (2014).
Baratta, M. G. et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proc. Natl Acad. Sci. USA 112, 232–237 (2015).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
Chen, S. et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260 (2015).
Johannessen, C. M. et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972 (2010).
Mann, K. M. et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl Acad. Sci. USA 109, 5934–5941 (2012).
Sos, M. L. et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc. Natl Acad. Sci. USA 109, 17034–17039 (2012).
Banerji, V. et al. The intersection of genetic and chemical genomic screens identifies GSK-3α as a target in human acute myeloid leukemia. J. Clin. Invest. 122, 935–947 (2012).
Gupte, A. et al. Systematic screening identifies dual PI3K and mTOR inhibition as a conserved therapeutic vulnerability in osteosarcoma. Clin. Cancer Res. 21, 3216–3229 (2015).
Heidorn, S. J. et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140, 209–221 (2010).
Hollestelle, A., Elstrodt, F., Nagel, J. H., Kallemeijn, W. W. & Schutte, M. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol. Cancer Res. 5, 195–201 (2007).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Punnoose, E. A. et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin. Cancer Res. 18, 2391–2401 (2012).
Iacovides, D. C. et al. Identification and quantification of AKT isoforms and phosphoforms in breast cancer using a novel nanofluidic immunoassay. Mol. Cell. Proteom. 12, 3210–3220 (2013).
Worboys, J. D., Sinclair, J., Yuan, Y. & Jorgensen, C. Systematic evaluation of quantotypic peptides for targeted analysis of the human kinome. Nat. Methods 11, 1041–1044 (2014).
Pe'er, D. & Hacohen, N. Principles and strategies for developing network models in cancer. Cell 144, 864–873 (2011).
Zaman, N. et al. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell Rep. 5, 216–223 (2013).
Hofree, M., Shen, J. P., Carter, H., Gross, A. & Ideker, T. Network-based stratification of tumor mutations. Nat. Methods 10, 1108–1115 (2013).
Lee, M. J. et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149, 780–794 (2012). This study used systems-level analysis to demonstrate rewiring of cell pathways regulating apoptosis upon sequential application of an EGFR inhibitor and the chemotherapeutic agent doxorubicin, leading to enhanced cell killing compared with simultaneous administration. It highlights how targeting cell signalling networks, rather than individual signalling proteins —the principle of 'network medicine' — can be an effective therapeutic strategy.
So, J. et al. Integrative analysis of kinase networks in TRAIL-induced apoptosis provides a source of potential targets for combination therapy. Sci. Signal. 8, rs3 (2015).
Chartier, M., Chenard, T., Barker, J. & Najmanovich, R. Kinome Render: a stand-alone and web-accessible tool to annotate the human protein kinome tree. PeerJ. 1, e126 (2013).
Perna, D. et al. BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc. Natl Acad. Sci. USA 112, E536–E545 (2015).
Smit, M. A. et al. ROCK1 is a potential combinatorial drug target for BRAF mutant melanoma. Mol. Syst. Biol. 10, 772 (2014).
Ahronian, L. G. et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 5, 358–367 (2015).
Nazarian, R. et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010).
Van Allen, E. M. et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 (2014).
Fedorenko, I. V. et al. Fibronectin induction abrogates the BRAF inhibitor response of BRAF V600E/PTEN-null melanoma cells. Oncogene http://dx.doi.org/10.1038/onc.2015.188 (2015).
Paraiso, K. H. et al. Ligand-independent EPHA2 signaling drives the adoption of a targeted therapy-mediated metastatic melanoma phenotype. Cancer Discov. 5, 264–273 (2015).
Straussman, R. et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 (2012).
Luo, J. et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848 (2009).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). References 111 and 112 demonstrate how synthetic-lethal functional screens can be used to identify potential therapeutic strategies for cancers expressing an oncogene challenging to conventional drug development pipelines, in this case oncogenic KRAS. In both cases a druggable protein kinase was identified as exhibiting a synthetic-lethal interaction with mutant KRAS, specifically polo-like kinase 1 (PLK1) and TANK-binding kinase 1 (TBK1).
Toyoshima, M. et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc. Natl Acad. Sci. USA 109, 9545–9550 (2012).
van der Meer, R., Song, H. Y., Park, S. H., Abdulkadir, S. A. & Roh, M. RNAi screen identifies a synthetic lethal interaction between PIM1 overexpression and PLK1 inhibition. Clin. Cancer Res. 20, 3211–3221 (2014).
Bhola, N. E. et al. Kinome-wide functional screen identifies role of PLK1 in hormone-independent, ER-positive breast cancer. Cancer Res. 75, 405–414 (2015).
Deng, T. et al. shRNA kinome screen identifies TBK1 as a therapeutic target for HER2+ breast cancer. Cancer Res. 74, 2119–2130 (2014).
Mendes-Pereira, A. M., Lord, C. J. & Ashworth, A. NLK is a novel therapeutic target for PTEN deficient tumour cells. PLoS ONE 7, e47249 (2012).
Bommi-Reddy, A. et al. Kinase requirements in human cells: III. Altered kinase requirements in VHL−/− cancer cells detected in a pilot synthetic lethal screen. Proc. Natl Acad. Sci. USA 105, 16484–16489 (2008).
Baldwin, A. et al. Kinase requirements in human cells: V. Synthetic lethal interactions between p53 and the protein kinases SGK2 and PAK3. Proc. Natl Acad. Sci. USA 107, 12463–12468 (2010).
Mohni, K. N., Kavanaugh, G. M. & Cortez, D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 74, 2835–2845 (2014).
Azorsa, D. O. et al. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. J. Transl Med. 7, 43 (2009).
Tibes, R. et al. RNAi screening of the kinome with cytarabine in leukemias. Blood 119, 2863–2872 (2012).
Josse, R. et al. ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase I inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 74, 6968–6979 (2014).
Possik, P. A. et al. Parallel in vivo and in vitro melanoma RNAi dropout screens reveal synthetic lethality between hypoxia and DNA damage response inhibition. Cell Rep. 9, 1375–1386 (2014).
Sullivan, K. D. et al. ATM and MET kinases are synthetic lethal with nongenotoxic activation of p53. Nat. Chem. Biol. 8, 646–654 (2012).
Lamba, S. et al. RAF suppression synergizes with MEK inhibition in KRAS mutant cancer cells. Cell Rep. 8, 1475–1483 (2014).
Chaudhuri, L. et al. CHK1 and WEE1 inhibition combine synergistically to enhance therapeutic efficacy in acute myeloid leukemia ex vivo. Haematologica 99, 688–696 (2014).
Milosevic, N. et al. Synthetic lethality screen identifies RPS6KA2 as modifier of epidermal growth factor receptor activity in pancreatic cancer. Neoplasia 15, 1354–1362 (2013).
Acknowledgements
R.J.D. is supported by a National Health and Medical Research Council (Australia) Principal Research Fellowship (APP1058540), J.W. is supported by a Cancer Council New South Wales grant (SRP11-01), and E.D.G.F. was supported by the Dutch Cancer Society for her postdoctoral position in Australia.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Supplementary information
41568_2016_BFnrc201518_MOESM234_ESM.pdf
Supplementary information S2 (figure) | Protein-protein interaction networks for driver kinases in different cancers. (PDF 699 kb)
Glossary
- PTEN
-
A phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase that antagonizes the PI3K signalling pathway and represents an important tumour suppressor in several human cancers.
- Reverse phase protein arrays
-
(RPPAs). Microarrays carrying large numbers of protein samples printed as individual spots, with detection of particular antigens achieved via incubation with a specific antibody.
- Breast cancer mRNA subtypes
-
Breast cancer can be subclassified into four major subtypes through gene expression profiling: luminal A, luminal B, HER2 and basal.
- Inositol polyphosphate 4-phosphatase type II
-
A phosphatidylinositol 3,4-bisphosphate 4-phosphatase that antagonizes the PI3K signalling pathway and represents a tumour suppressor.
- Liquid chromatography–tandem mass spectrometry
-
(LC–MS/MS). When applied to proteomics, the liquid chromatography fractionates the peptides present in a sample and the tandem MS determines peptide mass and then additional characteristics through fragmentation.
- Metal or metal oxide affinity chromatography
-
A technique used to purify phosphopeptides that exploits their binding to metal ions (such as iron) or metal oxides (such as titanium dioxide).
- Pseudokinase
-
A protein with a protein kinase-related domain that does not exhibit kinase activity owing to the absence of one or more conserved amino acid sequence motifs.
- NCI-60 cell line panel
-
A panel of 60 diverse human cancer cell lines used by the US National Cancer Institute to screen large numbers of chemical compounds, drugs and natural products for their biological activity.
- Sleeping Beauty transposon system
-
A technique that introduces a DNA vector at random sites throughout the mouse genome and thereby alters the expression of genes close to the insertion site.
Rights and permissions
About this article
Cite this article
Fleuren, E., Zhang, L., Wu, J. et al. The kinome 'at large' in cancer. Nat Rev Cancer 16, 83–98 (2016). https://doi.org/10.1038/nrc.2015.18
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc.2015.18
This article is cited by
-
Eph receptors and ephrins in cancer progression
Nature Reviews Cancer (2024)
-
Splicing alterations in pancreatic ductal adenocarcinoma: a new molecular landscape with translational potential
Journal of Experimental & Clinical Cancer Research (2023)
-
RSK3 switches cell fate: from stress-induced senescence to malignant progression
Journal of Experimental & Clinical Cancer Research (2023)
-
Biological and clinical implications of FGFR aberrations in paediatric and young adult cancers
Oncogene (2023)
-
Analysis and functional relevance of the chaperone TRAP-1 interactome in the metabolic regulation and mitochondrial integrity of cancer cells
Scientific Reports (2023)