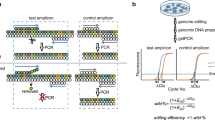
Abstract
Gene tagging with fluorescent proteins is essential for investigations of the dynamic properties of cellular proteins. CRISPR–Cas9 technology is a powerful tool for inserting fluorescent markers into all alleles of the gene of interest (GOI) and allows functionality and physiological expression of the fusion protein. It is essential to evaluate such genome-edited cell lines carefully in order to preclude off-target effects caused by (i) incorrect insertion of the fluorescent protein, (ii) perturbation of the fusion protein by the fluorescent proteins or (iii) nonspecific genomic DNA damage by CRISPR–Cas9. In this protocol, we provide a step-by-step description of our systematic pipeline to generate and validate homozygous fluorescent knock-in cell lines.
We have used the paired Cas9D10A nickase approach to efficiently insert tags into specific genomic loci via homology-directed repair (HDR) with minimal off-target effects. It is time-consuming and costly to perform whole-genome sequencing of each cell clone to check for spontaneous genetic variations occurring in mammalian cell lines. Therefore, we have developed an efficient validation pipeline of the generated cell lines consisting of junction PCR, Southern blotting analysis, Sanger sequencing, microscopy, western blotting analysis and live-cell imaging for cell-cycle dynamics. This protocol takes between 6 and 9 weeks. With this protocol, up to 70% of the targeted genes can be tagged homozygously with fluorescent proteins, thus resulting in physiological levels and phenotypically functional expression of the fusion proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Mahen, R. et al. Comparative assessment of fluorescent transgene methods for quantitative imaging in human cells. Mol. Biol. Cell 25, 3610–3618 (2014).
Otsuka, S. et al. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife 5, e19071 (2016).
Wachsmuth, M. et al. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 33, 384–389 (2015).
Politi, A.Z. et al. Quantitative mapping of endogenously fluorescently tagged proteins using FCS-calibrated four-dimensional imaging. Nat. Protoc. https://doi.org/10.1038/nprot.2018.040 (2018).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Trevino, A.E. et al. Genome editing using Cas9 nickases. Methods Enzymol. 546, 161–174 (2014).
Pattanayak, V. et al. Determining the specificities of TALENs, Cas9, and other genome-editing enzymes. Methods Enzymol. 546, 47–78 (2014).
Dambournet, D. et al. Tagging endogenous loci for live-cell fluorescence imaging and molecule counting using ZFNs, TALENs, and Cas9. Methods Enzymol. 546, 139–160 (2014).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Sander, J.D. & Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355 (2014).
Doudna, J.A. & Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science. 346, 1258096 (2014).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823 (2013).
Wang, H. et al. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264 (2016).
Sternberg, S.H. & Redding, S. DNA interrogation by the CRISPR. Nature 507, 62–67 (2014).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Shen, B. et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402 (2014).
Ran, F.A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Bothmer, A. et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 8, 13905 (2016).
Miyaoka, Y. et al. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci. Rep. 6, 23549 (2016).
Mao, Z. et al. DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia 11, 683–691 (2009).
Bertolini, L.R. et al. Increased gene targeting in Ku70 and Xrcc4 transiently deficient human somatic cells. Mol. Biotechnol. 41, 106–114 (2009).
Chu, V.T. et al. Increasing the efficiency of homology-directed repair for CrIsPr-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548 (2015).
Srivastava, M. et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 151, 1474–1487 (2012).
Boettcher, R. et al. Efficient chromosomal gene modification with CRISPR/Cas9 and PCR-based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res. 42, e89 (2014).
Maruyama, T. et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. FEBS J. 282, 4289–4294 (2015).
Greco, G.E. et al. SCR7 is neither a selective nor a potent inhibitor of human DNA ligase IV. DNA Repair (Amst) 43, 18–23 (2016).
Heijink, A.M. et al. The DNA damage response during mitosis. Mutat. Res. 750, 45–55 (2013).
Lin, S. et al. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014).
Branzei, D. & Foiani, M. Regulation of DNA repair throughout the cell cycle Nat. Rev. Mol. Cell Biol. 9, 297–308 (2008).
Mao, Z. et al. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 7, 2902–2906 (2008).
Pepperkok, R. & Ellenberg, J. High-throughput fluorescence microscopy for systems biology. Nat. Rev. Mol. Cell Biol. 7, 690–696 (2006).
Landry, J.J.M. et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3, 1213–1224 (2013).
Liang, X. et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 208, 44–53 (2015).
Kim, S. et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Zuris, J.A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Roberts, B. et al. Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Mol. Biol. Cell 28, 2854–2874 (2017).
Paix, A. et al. Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res. 44, e128 (2016).
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
Lemp, N.A. et al. Cryptic transcripts from a ubiquitous plasmid origin of replication confound tests for cis-regulatory function. Nucleic Acids Res. 40, 7280–7290 (2012).
Lukinaviius, G. et al. SiR–Hoechst is a far-red DNA stain for live-cell nanoscopy. Nat. Commun. 6, 8497 (2015).
Heller, C. Principles of DNA separation with capillary electrophoresis. Electrophoresis 22, 629–643 (2001).
Sanger, F., Nicklen, S. & Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467 (1977).
Brinkman, E.K. et al. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Hsu, P.D. et al. DNA targeting specificity of rNA-guided Cas9 nucleases. Nat. Biotechnol. 2, 827–832 (2013).
Doench, J.G. et al. Optimized sgrNA design to maximize activity and minimize off-target effects of CRISPR-cas9. Nat. Biotechnol. 34, 184–191 (2016).
Chen, X. et al. Fusion protein linkers: property, design and functionality. Adv. Drug Deliv. Rev. 65, 1357–1369 (2013).
Held, M. et al. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods 7, 747–754 (2010).
Acknowledgements
We thank the mechanical and electronics workshops of EMBL for custom hardware, the Advanced Light Microscopy Facility of EMBL for microscopy support and the Flow Cytometry Core Facility of EMBL for cell sorting. We gratefully acknowledge G. Reid for critical reading of the manuscript. This work was supported by grants to J.E. from the European Commission EU-FP7-Systems Microscopy NoE (grant agreement 258068), EU-FP7-MitoSys (grant agreement 241548) and iNEXT (grant agreement 653706), as well as by the European Molecular Biology Laboratory (B.K., B.N., M.K., Y.C., N.W. and J.E.). N.W. and Y.C. were supported by the EMBL International PhD Programme (EIPP).
Author information
Authors and Affiliations
Contributions
B.K. designed and performed the experiments. B.K. developed the protocol with the help of B.N., Y.C. and N.W. B.N. and M.K. tested the protocol. Y.C. and N.W. created the automation for the cell-cycle analysis. B.K. wrote the protocol with help from B.N., Y.C., N.W. and J.E. All authors contributed to the interpretation of the data and read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Donor plasmid.
The donor plasmid consists of the fluorescent marker gene (mEGFP in this case) which is flanked by 500-800 bp homology arms of the GOI. A linker is placed between the tag and the gene to maintain functionality of the tagged protein. As a backbone vector pUC-based plasmids are used.
Supplementary Figure 2 Scheme of expected junction PCR result.
A forward primer binding at the 5′ end outside of the left homology arm and a reverse primer binding to the fluorescent marker gene will result in one PCR product of the expected size. To test if all alleles are tagged with the fluorescent marker at the correct locus, a forward primer located 5′ outside of the left homology arm and a reverse primer 3′outside of the right homology arm were used. Two PCR products using this primer set will indicate heterozygote clones and one PCR product which runs at the expected size of the tagged gene will indicate homozygosity.
Supplementary Figure 3 Southern blot transfer of DNA.
This scheme depicts how to build up the southern blot transfer as described in step 48| Option B, step xii-xxiii). Two long filter papers are dipped into a tank filled with 10X SSC and used as wicks. Gel, nylon membrane and filter papers were sandwiched on a glass plate as depicted to transfer the DNA from the gel onto the membrane.
Supplementary Figure 4 Example sequences containing point mutations.
Sanger sequencing was performed with a PCR fragment (lower sequence line #1-278) which was aligned to the expected sequence of the GOI (middle sequence line labeled with #3201-3500). The overlaying green and red lines indicate a point mutation at position 3411 of the expected sequence, i.e. one allele has the nucleotide T at this position (green line) whereas another allele has the nucleotide A (red line) at the same position. This indicates a point mutation (red A) within one of the alleles.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 609 kb)
Rights and permissions
About this article
Cite this article
Koch, B., Nijmeijer, B., Kueblbeck, M. et al. Generation and validation of homozygous fluorescent knock-in cells using CRISPR–Cas9 genome editing. Nat Protoc 13, 1465–1487 (2018). https://doi.org/10.1038/nprot.2018.042
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.042
This article is cited by
-
StayGold variants for molecular fusion and membrane-targeting applications
Nature Methods (2024)
-
A nanobody-based strategy for rapid and scalable purification of human protein complexes
Nature Protocols (2024)
-
Oxidative Stress Induced by Arsenite is Involved in YTHDF2 Phase Separation
Biological Trace Element Research (2024)
-
A quantitative map of nuclear pore assembly reveals two distinct mechanisms
Nature (2023)
-
mRNA recognition and packaging by the human transcription–export complex
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.