Abstract
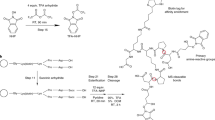
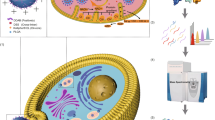
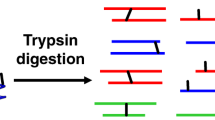
Many cellular functions necessitate structural assemblies of two or more associated proteins. The structural characterization of protein complexes using standard methods, such as X-ray crystallography, is challenging. Herein, we describe an orthogonal approach using hydrogen–deuterium-exchange mass spectrometry (HDXMS), cross-linking mass spectrometry (CXMS), and disulfide trapping to map interactions within protein complexes. HDXMS measures changes in solvent accessibility and hydrogen bonding upon complex formation; a decrease in HDX rate could account for newly formed intermolecular or intramolecular interactions. To distinguish between inter- and intramolecular interactions, we use a CXMS method to determine the position of direct interface regions by trapping intermolecular residues in close proximity to various cross-linkers (e.g., disuccinimidyl adipate (DSA)) of different lengths and reactive groups. Both MS-based experiments are performed on high-resolution mass spectrometers (e.g., an Orbitrap Elite hybrid mass spectrometer). The physiological relevance of the interactions identified through HDXMS and CXMS is investigated by transiently co-expressing cysteine mutant pairs, one mutant on each protein at the discovered interfaces, in an appropriate cell line, such as HEK293. Disulfide-trapped protein complexes are formed within cells spontaneously or are facilitated by addition of oxidation reagents such as H2O2 or diamide. Western blotting analysis, in the presence and absence of reducing reagents, is used to determine whether the disulfide bonds are formed in the proposed complex interface in physiologically relevant milieus. The procedure described here requires 1–2 months. We demonstrate this approach using the β2-adrenergic receptor-β-arrestin1 complex as the model system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Accession codes
References
Hartwell, L.H., Hopfield, J.J., Leibler, S. & Murray, A.W. From molecular to modular cell biology. Nature 402, C47–C52 (1999).
Rasmussen, S.G. et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Zheng, H. et al. X-ray crystallography over the past decade for novel drug discovery - where are we heading next? Expert Opin. Drug Discov. 10, 975–989 (2015).
Zhang, H. & Cramer, W.A. Problems in obtaining diffraction-quality crystals of hetero-oligomeric integral membrane proteins. J. Struct. Funct. Genomics 6, 219–223 (2005).
Slabinski, L. et al. The challenge of protein structure determination—lessons from structural genomics. Protein Sci. 16, 2472–2482 (2007).
Wlodawer, A., Minor, W., Dauter, Z. & Jaskolski, M. Protein crystallography for aspiring crystallographers or how to avoid pitfalls and traps in macromolecular structure determination. FEBS J. 280, 5705–5736 (2013).
Xiao, K., Chung, J. & Wall, A. The power of mass spectrometry in structural characterization of GPCR signaling. J. Recept. Signal Transduct. Res. 35, 213–219 (2015).
Kobilka, B. & Schertler, G.F. New G-protein-coupled receptor crystal structures: insights and limitations. Trends Pharmacol. Sci. 29, 79–83 (2008).
Karplus, M. & Petsko, G.A. Molecular dynamics simulations in biology. Nature 347, 631–639 (1990).
Sattler, M. & Fesik, S.W. Use of deuterium labeling in NMR: overcoming a sizeable problem. Structure 4, 1245–1249 (1996).
Wuthrich, K. The way to NMR structures of proteins. Nat. Struct. Biol. 8, 923–925 (2001).
Simon, B., Madl, T., Mackereth, C.D., Nilges, M. & Sattler, M. An efficient protocol for NMR-spectroscopy-based structure determination of protein complexes in solution. Angew. Chem. Int. Ed. Engl. 49, 1967–1970 (2010).
Wuthrich, K. Protein structure determination in solution by NMR spectroscopy. J. Biol. Chem. 265, 22059–22062 (1990).
Montalvao, R.W., Cavalli, A., Salvatella, X., Blundell, T.L. & Vendruscolo, M. Structure determination of protein-protein complexes using NMR chemical shifts: case of an endonuclease colicin-immunity protein complex. J. Am. Chem. Soc. 130, 15990–15996 (2008).
Jaroniec, C.P. Solid-state nuclear magnetic resonance structural studies of proteins using paramagnetic probes. Solid State Nucl. Magn. Reson. 43–44, 1–13 (2012).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Schmidt, R. et al. Spherical nanosized focal spot unravels the interior of cells. Nat. Methods 5, 539–544 (2008).
Kasas, S., Dumas, G., Dietler, G., Catsicas, S. & Adrian, M. Vitrification of cryoelectron microscopy specimens revealed by high-speed photographic imaging. J. Microsc. 211, 48–53 (2003).
Erni, R., Rossell, M.D., Kisielowski, C. & Dahmen, U. Atomic-resolution imaging with a sub-50-pm electron probe. Phys. Rev. Lett. 102, 096101 (2009).
Kaufmann, R., Muller, P., Hildenbrand, G., Hausmann, M. & Cremer, C. Analysis of Her2/neu membrane protein clusters in different types of breast cancer cells using localization microscopy. J. Microsc. 242, 46–54 (2011).
Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Rames, M., Yu, Y. & Ren, G. Optimized negative staining: a high-throughput protocol for examining small and asymmetric protein structure by electron microscopy. J. Vis. Exp. (90) e51087 http://dx.doi.org/10.3791/51087 (2014).
Booth, D.S., Avila-Sakar, A. & Cheng, Y. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J. Vis. Exp. http://dx.doi.org/10.3791/51087 (2011).
Shukla, A.K. et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222 (2014).
Roh, S.H. et al. Subunit conformational variation within individual GroEL oligomers resolved by cryo-EM. Proc. Natl. Acad. Sci. USA 114, 8259–8264 (2017).
Hite, R.K., Tao, X. & MacKinnon, R. Structural basis for gating the high-conductance Ca2+-activated K+ channel. Nature 541, 52–57 (2017).
Gao, Y. et al. Isolation and structure-function characterization of a signaling-active rhodopsin-G protein complex. J. Biol. Chem. 292, 14280–14289 (2017).
Guo, F. & Jiang, W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 1117, 401–443 (2014).
Adrian, M., Dubochet, J., Lepault, J. & McDowall, A.W. Cryo-electron microscopy of viruses. Nature 308, 32–36 (1984).
Dong, Y. et al. Antibody-induced uncoating of human rhinovirus B14. Proc. Natl. Acad. Sci. USA 114, 8017–8022 (2017).
Zhang, Y. et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546, 248–253 (2017).
Liang, Y.L. et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546, 118–123 (2017).
Beck, M. & Baumeister, W. Cryo-electron tomography: can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 26, 825–837 (2016).
Beck, M., Lucic, V., Forster, F., Baumeister, W. & Medalia, O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 449, 611–615 (2007).
Irobalieva, R.N., Martins, B. & Medalia, O. Cellular structural biology as revealed by cryo-electron tomography. J. Cell Sci. 129, 469–476 (2016).
Oikonomou, C.M. & Jensen, G.J. Cellular electron cryotomography: toward structural biology in situ. Annu. Rev. Biochem. 86, 873–896 (2017).
Liu, L., Boldon, L., Urquhart, M. & Wang, X. Small and wide angle X-ray scattering studies of biological macromolecules in solution. J. Vis. Exp. http://dx.doi.org/10.3791/4160 (2013).
Vachette, P., Koch, M.H. & Svergun, D.I. Looking behind the beamstop: X-ray solution scattering studies of structure and conformational changes of biological macromolecules. Methods Enzymol. 374, 584–615 (2003).
Hanneke, D., Fogwell, S. & Gabrielse, G. New measurement of the electron magnetic moment and the fine structure constant. Phys. Rev. Lett. 100, 120801 (2008).
Odom, B., Hanneke, D., D'Urso, B. & Gabrielse, G. New measurement of the electron magnetic moment using a one-electron quantum cyclotron. Phys. Rev. Lett. 97, 030801 (2006).
Altenbach, C., Flitsch, S.L., Khorana, H.G. & Hubbell, W.L. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry 28, 7806–7812 (1989).
Altenbach, C., Marti, T., Khorana, H.G. & Hubbell, W.L. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science 248, 1088–1092 (1990).
Elsasser, C., Brecht, M. & Bittl, R. Pulsed electron-electron double resonance on multinuclear metal clusters: assignment of spin projection factors based on the dipolar interaction. J. Am. Chem. Soc. 124, 12606–12611 (2002).
Endeward, B., Butterwick, J.A., MacKinnon, R. & Prisner, T.F. Pulsed electron-electron double-resonance determination of spin-label distances and orientations on the tetrameric potassium ion channel KcsA. J. Am. Chem. Soc. 131, 15246–15250 (2009).
Konermann, L., Vahidi, S. & Sowole, M.A. Mass spectrometry methods for studying structure and dynamics of biological macromolecules. Anal. Chem. 86, 213–232 (2014).
West, G.M. et al. Ligand-dependent perturbation of the conformational ensemble for the GPCR beta2 adrenergic receptor revealed by HDX. Structure 19, 1424–1432 (2011).
Lodowski, D.T., Palczewski, K. & Miyagi, M. Conformational changes in the g protein-coupled receptor rhodopsin revealed by histidine hydrogen-deuterium exchange. Biochemistry 49, 9425–9427 (2010).
Zhou, M. & Robinson, C.V. Flexible membrane proteins: functional dynamics captured by mass spectrometry. Curr. Opin. Struct. Biol. 28, 122–130 (2014).
Ishii, K., Noda, M. & Uchiyama, S. Mass spectrometric analysis of protein-ligand interactions. Biophys. Physicobiol. 13, 87–95 (2016).
Kang, Y. et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 (2015).
Liko, I., Allison, T.M., Hopper, J.T. & Robinson, C.V. Mass spectrometry guided structural biology. Curr. Opin. Struct. Biol. 40, 136–144 (2016).
Artigues, A. et al. Protein structural analysis via mass spectrometry-based proteomics. Adv. Exp. Med. Biol. 919, 397–431 (2016).
Petrotchenko, E.V. & Borchers, C.H. Modern mass spectrometry-based structural proteomics. Adv. Protein Chem. Struct. Biol. 95, 193–213 (2014).
Serpa, J.J. et al. Mass spectrometry-based structural proteomics. Eur. J. Mass Spectrom. 18, 251–267 (2012).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016).
Walzthoeni, T., Leitner, A., Stengel, F. & Aebersold, R. Mass spectrometry supported determination of protein complex structure. Curr. Opin. Struct. Biol. 23, 252–260 (2013).
Ghanouni, P. et al. Functionally different agonists induce distinct conformations in the G protein coupling domain of the beta 2 adrenergic receptor. J. Biol. Chem. 276, 24433–24436 (2001).
Ghanouni, P., Steenhuis, J.J., Farrens, D.L. & Kobilka, B.K. Agonist-induced conformational changes in the G-protein-coupling domain of the beta 2 adrenergic receptor. Proc. Natl. Acad. Sci. USA 98, 5997–6002 (2001).
Reiner, S., Ambrosio, M., Hoffmann, C. & Lohse, M.J. Differential signaling of the endogenous agonists at the beta2-adrenergic receptor. J. Biol. Chem. 285, 36188–36198 (2010).
Swaminath, G. et al. Probing the beta2 adrenoceptor binding site with catechol reveals differences in binding and activation by agonists and partial agonists. J. Biol. Chem. 280, 22165–22171 (2005).
Yao, X. et al. Coupling ligand structure to specific conformational switches in the beta2-adrenoceptor. Nat. Chem. Biol. 2, 417–422 (2006).
Bokoch, M.P. et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463, 108–112 (2010).
Nygaard, R. et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell 152, 532–542 (2013).
Li, S., Lee, S.Y. & Chung, K.Y. Conformational analysis of G protein-coupled receptor signaling by hydrogen/deuterium exchange mass spectrometry. Methods Enzymol. 557, 261–278 (2015).
Kahsai, A.W. et al. Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nat. Chem. Biol. 7, 692–700 (2011).
Kahsai, A.W., Rajagopal, S., Sun, J. & Xiao, K. Monitoring protein conformational changes and dynamics using stable-isotope labeling and mass spectrometry. Nat. Protoc. 9, 1301–1319 (2014).
Xiao, K. et al. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc. Natl. Acad. Sci. USA 104, 12011–12016 (2007).
Xiao, K. & Shenoy, S.K. Beta2-adrenergic receptor lysosomal trafficking is regulated by ubiquitination of lysyl residues in two distinct receptor domains. J. Biol. Chem. 286, 12785–12795 (2011).
Xiao, K., Shenoy, S.K., Nobles, K. & Lefkowitz, R.J. Activation-dependent conformational changes in {beta}-arrestin 2. J. Biol. Chem. 279, 55744–55753 (2004).
Xiao, K. et al. Global phosphorylation analysis of beta-arrestin-mediated signaling downstream of a seven transmembrane receptor (7TMR). Proc. Natl. Acad. Sci. USA 107, 15299–15304 (2010).
Xie, L. et al. Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci. Signal. 2, ra33 (2009).
Nobles, K.N., Guan, Z., Xiao, K., Oas, T.G. & Lefkowitz, R.J. The active conformation of beta-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of beta-arrestins1 and -2. J. Biol. Chem. 282, 21370–21381 (2007).
Nobles, K.N. et al. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci. Signal. 4, ra51 (2011).
Petrotchenko, E.V. et al. BiPS, a photocleavable, isotopically coded, fluorescent cross-linker for structural proteomics. Mol. Cell. Proteomics 8, 273–286 (2009).
Englander, J.J. et al. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc. Natl. Acad. Sci. USA 100, 7057–7062 (2003).
Englander, S.W. Hydrogen exchange and mass spectrometry: a historical perspective. J. Am. Soc. Mass Spectrom. 17, 1481–1489 (2006).
Busenlehner, L.S. & Armstrong, R.N. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Arch. Biochem. Biophys. 433, 34–46 (2005).
Wales, T.E. & Engen, J.R. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006).
Maier, C.S. & Deinzer, M.L. Protein conformations, interactions, and H/D exchange. Methods Enzymol. 402, 312–360 (2005).
Zhang, Z. & Smith, D.L. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993).
Konermann, L., Pan, J. & Liu, Y.H. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011).
Chalmers, M.J., Busby, S.A., Pascal, B.D., West, G.M. & Griffin, P.R. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev. Proteomics 8, 43–59 (2011).
Kan, Z.Y., Walters, B.T., Mayne, L. & Englander, S.W. Protein hydrogen exchange at residue resolution by proteolytic fragmentation mass spectrometry analysis. Proc. Natl. Acad. Sci. USA 110, 16438–16443 (2013).
Leitner, A., Faini, M., Stengel, F. & Aebersold, R. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 (2016).
Rappsilber, J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540 (2011).
Dobson, C.L. et al. Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo. Sci. Rep. 6, 38644 (2016).
Bhat, J.Y. et al. Mechanism of enzyme repair by the AAA+ chaperone rubisco activase. Mol. Cell 67, 744.e6–756.e6 (2017).
Rozbesky, D. et al. Chemical cross-linking and H/D exchange for fast refinement of protein crystal structure. Anal. Chem. 84, 867–870 (2012).
Haladova, K. et al. The combination of hydrogen/deuterium exchange or chemical cross-linking techniques with mass spectrometry: mapping of human 14-3-3zeta homodimer interface. J. Struct. Biol. 179, 10–17 (2012).
Mendoza, V.L. & Vachet, R.W. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 28, 785–815 (2009).
McKee, C.J., Kessl, J.J., Norris, J.O., Shkriabai, N. & Kvaratskhelia, M. Mass spectrometry-based footprinting of protein-protein interactions. Methods 47, 304–307 (2009).
Liu, H. et al. Mass spectrometry-based footprinting reveals structural dynamics of loop E of the chlorophyll-binding protein CP43 during photosystem II assembly in the Cyanobacterium Synechocystis 6803. J. Biol. Chem. 288, 14212–14220 (2013).
Kobilka, B.K. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal. Biochem. 231, 269–271 (1995).
Rasmussen, S.G. et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469, 175–180 (2011).
Ahn, J., Jung, M.C., Wyndham, K., Yu, Y.Q. & Engen, J.R. Pepsin immobilized on high-strength hybrid particles for continuous flow online digestion at 10,000 psi. Anal. Chem. 84, 7256–7262 (2012).
Belov, M.E. et al. Automated gain control and internal calibration with external ion accumulation capillary liquid chromatography-electrospray ionization Fourier transform ion cyclotron resonance. Anal. Chem. 75, 4195–4205 (2003).
Wales, T.E., Eggertson, M.J. & Engen, J.R. Considerations in the analysis of hydrogen exchange mass spectrometry data. Methods Mol. Biol. 1007, 263–288 (2013).
Kazazic, S. et al. Automated data reduction for hydrogen/deuterium exchange experiments, enabled by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 21, 550–558 (2010).
Rand, K.D., Adams, C.M., Zubarev, R.A. & Jorgensen, T.J. Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. J. Am. Chem. Soc. 130, 1341–1349 (2008).
Zehl, M., Rand, K.D., Jensen, O.N. & Jorgensen, T.J. Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. J. Am. Chem. Soc. 130, 17453–17459 (2008).
Zhang, X. et al. Dynamics of the beta2-adrenergic G-protein coupled receptor revealed by hydrogen-deuterium exchange. Anal. Chem. 82, 1100–1108 (2010).
Haas, W. et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics 5, 1326–1337 (2006).
Thevenon-Emeric, G., Kozlowski, J., Zhang, Z. & Smith, D.L. Determination of amide hydrogen exchange rates in peptides by mass spectrometry. Anal. Chem. 64, 2456–2458 (1992).
Keppel, T.R., Howard, B.A. & Weis, D.D. Mapping unstructured regions and synergistic folding in intrinsically disordered proteins with amide H/D exchange mass spectrometry. Biochemistry 50, 8722–8732 (2011).
Bai, Y., Milne, J.S., Mayne, L. & Englander, S.W. Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 (1993).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J.V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Kang, Y. et al. A structural snapshot of the rhodopsin-arrestin complex. FEBS J. 283, 816–821 (2016).
Bui, K.H. et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243 (2013).
Tosi, A. et al. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154, 1207–1219 (2013).
Murakami, K. et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342, 1238724 (2013).
Cormann, K.U., Moller, M. & Nowaczyk, M.M. Critical assessment of protein cross-linking and molecular docking: an updated model for the interaction between photosystem II and Psb27. Front. Plant Sci. 7, 157 (2016).
Powell, C.J. et al. Dissecting the molecular assembly of the Toxoplasma gondii MyoA motility complex. J. Biol. Chem. 292, 19469–19477 (2017).
Stjepanovic, G. et al. Assembly and dynamics of the autophagy-initiating Atg1 complex. Proc. Natl. Acad. Sci. USA 111, 12793–12798 (2014).
Di Michele, M. et al. Limited proteolysis combined with stable isotope labeling reveals conformational changes in protein (pseudo)kinases upon binding small molecules. J. Proteome Res. 14, 4179–4193 (2015).
O'Brien, D.P. et al. SEC-SAXS and HDX-MS: a powerful combination. The case of the calcium-binding domain of a bacterial toxin. Biotechnol. Appl. Biochem. 65, 62–68 (2017).
He, W. et al. Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proc. Natl. Acad. Sci. USA 108, 12307–12312 (2011).
Tian, S. et al. Pih1p-Tah1p puts a lid on hexameric AAA+ ATPases Rvb1/2p. Structure 25, 1519.e4–1529.e4 (2017).
Hamuro, Y. et al. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J. Biomol. Tech. 14, 171–182 (2003).
Horn, J.R. et al. The role of protein dynamics in increasing binding affinity for an engineered protein-protein interaction established by H/D exchange mass spectrometry. Biochemistry 45, 8488–8498 (2006).
Pascal, B.D. et al. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 (2012).
Guttman, M., Weis, D.D., Engen, J.R. & Lee, K.K. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J. Am. Soc. Mass Spectrom. 24, 1906–1912 (2013).
Kan, Z.Y., Mayne, L., Chetty, P.S. & Englander, S.W. ExMS: data analysis for HX-MS experiments. J. Am. Soc. Mass Spectrom. 22, 1906–1915 (2011).
Lindner, R. et al. Hexicon 2: automated processing of hydrogen-deuterium exchange mass spectrometry data with improved deuteration distribution estimation. J. Am. Soc. Mass Spectrom. 25, 1018–1028 (2014).
Lou, X. et al. Deuteration distribution estimation with improved sequence coverage for HX/MS experiments. Bioinformatics 26, 1535–1541 (2010).
Rey, M. et al. Mass spec studio for integrative structural biology. Structure 22, 1538–1548 (2014).
Wei, H. et al. Using hydrogen/deuterium exchange mass spectrometry to study conformational changes in granulocyte colony stimulating factor upon PEGylation. J. Am. Soc. Mass Spectrom. 23, 498–504 (2012).
Acknowledgements
We thank R.J. Lefkowitz (Duke University) and B.K. Kobilka (Stanford University) for invaluable guidance and enthusiastic support, and A.K. Shukla and A.W. Kahsai for stimulating ideas. This work was supported, in part, by US National Institutes of Health grant HL-075443 Proteomics Core support to K.X. This publication was also made possible by seed funding support to K.X. from the Department of Pharmacology and Chemical Biology, the University of Pittsburgh, the Vascular Medicine Institute, the Hemophilia Center of Western Pennsylvania, and the Institute for Transfusion Medicine.
Author information
Authors and Affiliations
Contributions
K.X. conceived the orthogonal approach combining HDXMS, CXMS, and disulfide trapping, and designed the experiments; S.L. conducted the HDXMS experiments; J.Q. conducted the CXMS experiments; M.C. conducted the disulfide-trapping experiments; K.X., Y.Z., H.L., A.B., T.J.C., X.L., Y.X., L.J.C., and S.L. analyzed data and wrote the paper; all authors read, edited, and discussed the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Xiao, K., Zhao, Y., Choi, M. et al. Revealing the architecture of protein complexes by an orthogonal approach combining HDXMS, CXMS, and disulfide trapping. Nat Protoc 13, 1403–1428 (2018). https://doi.org/10.1038/nprot.2018.037
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.037
This article is cited by
-
Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition
Nature Communications (2022)
-
Detection of structural and conformational changes in ALS-causing mutant profilin-1 with hydrogen/deuterium exchange mass spectrometry and bioinformatics techniques
Metabolic Brain Disease (2022)
-
A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.