Abstract
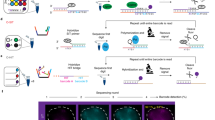
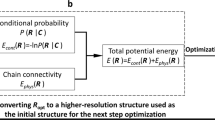
Fluorescence imaging and chromosome conformation capture assays such as Hi-C are key tools for studying genome organization. However, traditionally, they have been carried out independently, making integration of the two types of data difficult to perform. By trapping individual cell nuclei inside a well of a 384-well glass-bottom plate with an agarose pad, we have established a protocol that allows both fluorescence imaging and Hi-C processing to be carried out on the same single cell. The protocol identifies 30,000–100,000 chromosome contacts per single haploid genome in parallel with fluorescence images. Contacts can be used to calculate intact genome structures to better than 100-kb resolution, which can then be directly compared with the images. Preparation of 20 single-cell Hi-C libraries using this protocol takes 5 d of bench work by researchers experienced in molecular biology techniques. Image acquisition and analysis require basic understanding of fluorescence microscopy, and some bioinformatics knowledge is required to run the sequence-processing tools described here.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Cremer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292–301 (2001).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Dixon, J.R. et al. Topological domains in mammalian genomes dentified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Rao, S.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Nora, E.P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Bystricky, K., Heun, P., Gehlen, L., Langowski, J. & Gasser, S.M. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. USA 101, 16495–16500 (2004).
Nagano, T. et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat. Protoc. 10, 1986–2003 (2015).
Ramani, V. et al. Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017).
Flyamer, I.M. et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 (2017).
Stevens, T.J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Research 4, 1310 (2015).
Wlodkowic, D. & Darzynkiewicz, Z. Please do not disturb: destruction of chromatin structure by supravital nucleic acid probes revealed by a novel assay of DNA-histone interaction. Cytometry A 73, 877–879 (2008).
Freimann, R., Kramer, S., Böhmler, A. & Wutz, A. Gewinnung haploider Stammzellkulturen der Maus für genetische screens. BIOspektrum 20, 416–418 (2014).
Krishan, A. et al. Flow immunocytochemistry of marker expression in cells from body cavity fluids. Cytometry A 77, 132–143 (2010).
Leeb, M. & Wutz, A. Derivation of haploid embryonic stem cells from mouse embryos. Nature 479, 131–134 (2011).
Naumova, N. et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013).
Salzman, G.C., Wilder, M.E. & Jett, J.H. Light scattering with stream-in-air flow systems. J. Histochem. Cytochem. 27, 264–267 (1979).
Riddell, A., Gardner, R., Perez-Gonzalez, A., Lopes, T. & Martinez, L. Rmax: a systematic approach to evaluate instrument sort performance using center stream catch. Methods 82, 64–73 (2015).
Acknowledgements
We thank the Cancer Research UK (CRUK) Cambridge Institute for DNA sequencing. This work was supported by the Wellcome Trust (206291/Z/17/Z and 082010/Z/07/Z), the EC FP7 4DCellFate project (277899) and the MRC (MR/P019471/1, MR/M010082/1).
Author information
Authors and Affiliations
Contributions
D.L. and S.B. developed the experimental part of the protocol with assistance from Y.C. A.R. developed the flow cytometry methods to sort haploid-cell nuclei. T.J.S. and W.B. wrote the data-processing pipeline with assistance from L.P.A. and K.J.W. S.F.L. and D.K. helped to implement microscopy into the protocol. M.L. and B.H. provided expertise in haploid embryonic stem cell culture. E.D.L. supervised the project. D.L., T.J.S. and E.D.L. wrote the manuscript with contributions from all the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Lando, D., Basu, S., Stevens, T. et al. Combining fluorescence imaging with Hi-C to study 3D genome architecture of the same single cell. Nat Protoc 13, 1034–1061 (2018). https://doi.org/10.1038/nprot.2018.017
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.017
This article is cited by
-
Live-cell three-dimensional single-molecule tracking reveals modulation of enhancer dynamics by NuRD
Nature Structural & Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.