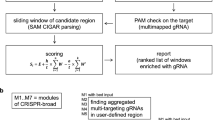
Abstract
CRISPR (clustered regularly interspaced short palindromic repeats) genome-editing experiments offer enormous potential for the evaluation of genomic loci using arrayed single guide RNAs (sgRNAs) or pooled sgRNA libraries. Numerous computational tools are available to help design sgRNAs with optimal on-target efficiency and minimal off-target potential. In addition, computational tools have been developed to analyze deep-sequencing data resulting from genome-editing experiments. However, these tools are typically developed in isolation and oftentimes are not readily translatable into laboratory-based experiments. Here, we present a protocol that describes in detail both the computational and benchtop implementation of an arrayed and/or pooled CRISPR genome-editing experiment. This protocol provides instructions for sgRNA design with CRISPOR (computational tool for the design, evaluation, and cloning of sgRNA sequences), experimental implementation, and analysis of the resulting high-throughput sequencing data with CRISPResso (computational tool for analysis of genome-editing outcomes from deep-sequencing data). This protocol allows for design and execution of arrayed and pooled CRISPR experiments in 4–5 weeks by non-experts, as well as computational data analysis that can be performed in 1–2 d by both computational and noncomputational biologists alike using web-based and/or command-line versions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–971 (2015).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Canver, M.C. et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 289, 21312–21324 (2014).
Ran, F.A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Hsu, P.D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016).
Pinello, L. et al. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 34, 695–697 (2016).
Canver, M.C. et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015).
Canver, M.C. et al. Variant-aware saturating mutagenesis using multiple Cas9 nucleases identifies regulatory elements at trait-associated loci. Nat. Genet. 49, 625–634 (2017).
Canver, M.C., Bauer, D.E. & Orkin, S.H. Functional interrogation of non-coding DNA through CRISPR genome editing. Methods 121–122, 118–129 (2017).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Koike-Yusa, H., Li, Y., Tan, E.-P., Velasco-Herrera, M.D.C. & Yusa, K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 32, 267–273 (2014).
Zhou, Y. et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491 (2014).
Gilbert, L.A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Sanjana, N.E. et al. High-resolution interrogation of functional elements in the noncoding genome. Science 353, 1545–1549 (2016).
Sanjana, N.E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Doench, J.G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Park, J., Kim, J. & Bae, S. Cas-Database: web-based genome-wide guide RNA library design for gene knockout screens using CRISPR-Cas9. Bioinformatics 32, 2017–2023 (2016).
Bae, S., Park, J. & Kim, J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Xiao, A. et al. CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30, 1180–1182 (2014).
Stemmer, M., Thumberger, T., Del Sol Keyer, M., Wittbrodt, J. & Mateo, J.L. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10, 1–11 (2015).
Cradick, T.J., Qiu, P., Lee, C.M., Fine, E.J. & Bao, G. COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol. Ther. Nucleic Acids 3, e214 (2014).
Montague, T.G., Cruz, J.M., Gagnon, J.A., Church, G.M. & Valen, E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, 401–407 (2014).
Labun, K., Montague, T.G., Gagnon, J.A., Thyme, S.B. & Valen, E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276 (2016).
Naito, Y., Hino, K., Bono, H. & Ui-Tei, K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 (2015).
Ma, J. et al. CRISPR-DO for genome-wide CRISPR design and optimization. Bioinformatics 32, 3336–3338 (2016).
Liu, H. et al. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics 31, 3676–3678 (2015).
Lei, Y. et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496 (2014).
Singh, R., Kuscu, C., Quinlan, A., Qi, Y. & Adli, M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 43, e118 (2015).
Heigwer, F., Kerr, G. & Boutros, M. E-CRISP: fast CRISPR target site identification. Nat. Methods 11, 122–123 (2014).
Gratz, S.J. et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014).
Meier, J.A., Zhang, F. & Sanjana, N. GUIDES: sgRNA design for loss-of-function screens. Nat. Methods 14, 831–832 (2017).
Perez, A.R. et al. GuideScan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 35, 347–349 (2017).
O'Brien, A. & Bailey, T.L. GT-Scan: identifying unique genomic targets. Bioinformatics 30, 2673–2675 (2014).
Wong, N., Liu, W. & Wang, X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218 (2015).
Zhu, L.J., Holmes, B.R., Aronin, N. & Brodsky, M.H. CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS One 9, e108424 (2014).
Xie, S., Shen, B., Zhang, C., Huang, X. & Zhang, Y. SgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One 9, e100448 (2014).
Prykhozhij, S.V., Rajan, V., Gaston, D. & Berman, J.N. CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS One 10, e0119372 (2015).
Tycko, J., Myer, V.E. & Hsu, P.D. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol. Cell 63, 355–370 (2016).
Fusi, N., Smith, I., Doench, J. & Listgarten, J. In silico predictive modeling of CRISPR/Cas9 guide efficiency. Preprint at bioRxiv, doi.org/10.1101/021568 (2015).
Chari, R., Mali, P., Moosburner, M. & Church, G.M. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods 12, 823–826 (2015).
Xu, H. et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25, 1147–1157 (2015).
Doench, J. et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014).
Wang, T., Wei, J.J., Sabatini, D.M. & Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014).
Moreno-Mateos, M.A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015).
Housden, B.E. et al. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci. Signal. 8, rs9 (2015).
Ren, X. et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 9, 1151–1162 (2014).
Farboud, B. & Meyer, B.J. Dramatic enhancement of genome editing by CRISPR/cas9 through improved guide RNA design. Genetics 199, 959–971 (2015).
Bae, S., Kweon, J., Kim, H.S. & Kim, J.-S. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11, 705–706 (2014).
Güell, M., Yang, L. & Church, G.M. Genome editing assessment using CRISPR genome analyzer (CRISPR-GA). Bioinformatics 30, 2968–2970 (2014).
Park, J., Lim, K., Kim, J.-S. & Bae, S. Cas-Analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288 (2017).
Xue, L.J. & Tsai, C.J. AGEseq: analysis of genome editing by sequencing. Mol. Plant 8, 1428–1430 (2015).
Lindsay, H. et al. CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol. 34, 701–702 (2016).
Boel, A. et al. BATCH-GE: batch analysis of next-generation sequencing data for genome editing assessment. Sci. Rep. 6, 30330 (2016).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Nelson, C.E. & Gersbach, C.A. Engineering delivery vehicles for genome editing. Annu. Rev. Chem. Biomol. Eng. 7, 637–662 (2016).
Yin, H., Kauffman, K.J. & Anderson, D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 16, 387–399 (2017).
Montalbano, A., Canver, M.C. & Sanjana, N.E. High-throughput approaches to pinpoint function within the noncoding genome. Mol. Cell 68, 44–59 (2017).
Tsai, S.Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Frock, R.L. et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 33, 179–186 (2015).
Yan, W.X. et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 8, 15058 (2017).
Tsai, S.Q. et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
Park, J. et al. Digenome-seq web tool for profiling CRISPR specificity. Nat. Methods 14, 548–549 (2017).
Cameron, P. et al. Mapping the genomic landscape of CRISPR–Cas9 cleavage. Nat. Methods 14, 600–606 (2017).
Shi, J. et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Cheng, A.W. et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163–1171 (2013).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Horlbeck, M.A. et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5, e19760 (2016).
Horlbeck, M.A. et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife 5, e12677 (2016).
Liu, S.J. et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, aah7111 (2017).
Joung, J. et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548, 343–346 (2017).
Kleinstiver, B.P. et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Slaymaker, I.M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Chen, J.S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017).
JoVE Science Education Database. Basic Methods in Cellular and Molecular Biology. Gel Purification. JoVE, Cambridge, MA. https://www.jove.com/science-education/5063/gel-purification (2018).
Froger, A. & Hall, J.E. Transformation of plasmid DNA into E. coli using the heat shock method. J. Vis. Exp. e253 (6) http://dx.doi.org/10.3791/253 (2007)
JoVE Science Education Database. Basic Methods in Cellular and Molecular Biology. Bacterial Transformation: The Heat Shock Method. JoVE, Cambridge, MA. https://www.jove.com/science-education/5059/bacterial-transformation-the-heat-shock-method (2018).
Kutner, R.H., Zhang, X.-Y. & Reiser, J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 4, 495–505 (2009).
Coufal, N.G. et al. L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009).
Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8, 186–194 (1998).
Bolger, A.M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Ellis, E.L. & Delbrück, M. The growth of bacteriophage. J. Gen. Physiol. 22, 365–384 (1939).
Stent, G. Molecular Biology of Bacterial Viruses (Freeman, 1963).
Choi, C., Kuatsjah, E., Wu, E. & Yuan, S. The effect of cell size on the burst size of T4 bacteriophage infections of Escherichia coli B23. J. Exp. Microbiol. Immunol. 14, 85–91 (2010).
Brendel, C. & Williams, D.A. Unexpected help: mTOR meets lentiviral vectors. Blood 124, 832–833 (2014).
O'Doherty, U., Swiggard, W.J. & Malim, M.H. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74, 10074–10080 (2000).
Sims, D. et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome Biol. 12, R104 (2011).
Acknowledgements
M.C.C. was supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Award (F30DK103359). M.H. was funded by National Institutes of Health (NIH)/National Human Genome Research Institute (NHGRI) grant 5U41HG002371-15 and NIH/National Cancer Institute (NCI) grant 5U54HG007990-02 and by a grant from the California Institute of Regenerative Medicine, CIRM GC1R-06673C. D.E.B. was supported by the National Heart, Lung, and Blood Institute (NHLBI) (DP2OD022716, P01HL032262), the Burroughs Wellcome Fund, and a Doris Duke Charitable Foundation Innovations in Clinical Research Award. S.H.O. was supported by an award from the NHLBI (P01HL032262) and an award from the NIDDK (P30DK049216, Center of Excellence in Molecular Hematology). N.E.S. was supported by the NIH through the NHGRI (R00-HG008171). L.P. was supported by an NHGRI Career Development Award (R00HG008399) and the Defense Advanced Research Projects Agency (HR0011-17-2-0042).
Author information
Authors and Affiliations
Contributions
M.C.C., M.H., and L.P. conceived this project. M.H. and J.-P.C. created CRISPOR. L.P., M.C.C., D.E.B., and G.-C.Y. created CRISPResso. M.C.C. and D.E.B. performed the experiments. M.C.C., D.E.B., S.H.O., N.E.S., O.S., G.-C.Y., F.Z., and L.P. analyzed the experimental data. M.C.C., M.H., and L.P. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Screenshot of sharing disk volumes and allocating memory with Docker containers.
a, Screenshot using Docker to ensure that the drive(s) you want to be available to the container is/are checked (under Settings…/ Shared Drives). b, Screenshot using Docker to allocate enough memory to the container (under Settings…/ Advanced).
Supplementary Figure 2 Pooled sgRNA library preparation and analysis.
a, Representative results for lsPCR1. b, Representative results for lsPCR2. c, Gradient PCR for lentiGuide-Puro-specific primers or locus-specific primers for laPCR1. d, Representative results for laPCR2.
Supplementary Figure 3 Example amplicon and regions description files.
a, Example of a properly formatted amplicon description file. This file is a tab delimited text file with up to 5 columns (first 2 columns required). No column heading is required. b, Example of a properly formatted regions description file. This file is a tab delimited text file with up to 7 columns (4 required) and contains the coordinates of the regions to analyze and some additional information. No column heading is required.
Supplementary Figure 4 Visualization of the distribution of identified alleles generated from targeting BCL11A exon 2.
Nucleotides are indicated by unique colors (A = green; C = red; G = yellow; T = purple). Substitutions are shown in bold font. Red rectangles highlight inserted sequences. Horizontal dashed lines indicate deleted sequences. The vertical dashed line indicates the predicted double-strand break position.
Supplementary Figure 5 Direct comparison of BCL11A exon 2 sequence between a BCL11A exon 2 targeted sgRNA sample (“edited”) and a non-edited control sample (“non-edited”).
a, Distribution of editing outcomes (unmodified, NHEJ, HDR, and mixed alleles) for treated (edited) and control (non- edited) samples. b, Comparison of the percent different editing outcomes (unmodified, NHEJ, HDR, and mixed alleles) between the treated (edited) and control (non-edited) samples. c, Combined (substitutions/deletions/insertions) mutation position distribution for treated (edited) and control (non- edited) samples. The vertical dashed line indicates the position of predicted Cas9 cleavage. The position of the sgRNA is shown in gray. d, Comparison of the percent different combined mutations (substitutions/deletions/insertions) between the treated (edited) and control (non-edited) samples. The vertical dashed line indicates the position of predicted Cas9 cleavage. The position of the sgRNA is shown in gray.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–8. (PDF 1462 kb)
Rights and permissions
About this article
Cite this article
Canver, M., Haeussler, M., Bauer, D. et al. Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments. Nat Protoc 13, 946–986 (2018). https://doi.org/10.1038/nprot.2018.005
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.005
This article is cited by
-
Aggresome formation promotes ASK1/JNK signaling activation and stemness maintenance in ovarian cancer
Nature Communications (2024)
-
Base editor scanning charts the DNMT3A activity landscape
Nature Chemical Biology (2023)
-
A pan-CRISPR analysis of mammalian cell specificity identifies ultra-compact sgRNA subsets for genome-scale experiments
Nature Communications (2022)
-
CRISPR-assisted rational flux-tuning and arrayed CRISPRi screening of an l-proline exporter for l-proline hyperproduction
Nature Communications (2022)
-
Tutorial: design and execution of CRISPR in vivo screens
Nature Protocols (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.