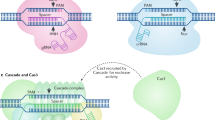
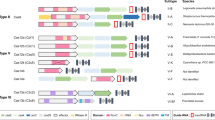
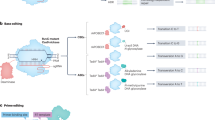
Abstract
Cpf1, a CRISPR endonuclease discovered in Prevotella and Francisella 1 bacteria, offers an alternative platform for CRISPR-based genome editing beyond the commonly used CRISPR–Cas9 system originally discovered in Streptococcus pyogenes. This protocol enables the design of engineered CRISPR–Cpf1 components, both CRISPR RNAs (crRNAs) to guide the endonuclease and Cpf1 mRNAs to express the endonuclease protein, and provides experimental procedures for effective genome editing using this system. We also describe quantification of genome-editing activity and off-target effects of the engineered CRISPR–Cpf1 in human cell lines using both T7 endonuclease I (T7E1) assay and targeted deep sequencing. This protocol enables rapid construction and identification of engineered crRNAs and Cpf1 mRNAs to enhance genome-editing efficiency using the CRISPR–Cpf1 system, as well as assessment of target specificity within 2 months. This protocol may also be appropriate for fine-tuning other types of CRISPR systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Zetsche, B. et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34 (2017).
Kim, Y. et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol. 34, 808–810 (2016).
Zhang, Y. et al. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv. 3, e1602814 (2017).
Watkins-Chow, D.E. et al. Highly-efficient Cpf1-mediated gene targeting in mice following high concentration pronuclear injection. G3: Genes Genomes Genet. 7, 719–722 (2016).
Endo, A., Masafumi, M., Kaya, H. & Toki, S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 6, 38169 (2016).
Kim, H. et al. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8, 14406 (2017).
Tang, X. et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 3, 17018 (2017).
Wang, M., Mao, Y., Lu, Y., Tao, X. & Zhu, J.K. Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol. Plant 10, 1011–1013 (2017).
Xu, R. et al. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 15, 713–717 (2017).
Yin, X. et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745–757 (2017).
Hur, J.K. et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol. 34, 807–808 (2016).
Gao, L. et al. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 35, 789–792 (2017).
Fonfara, I., Richter, H., Bratovic, M., Le Rhun, A. & Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016).
Yamano, T. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016).
Dong, D. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522–526 (2016).
Gao, P., Yang, H., Rajashankar, K.R., Huang, Z. & Patel, D.J. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 26, 901–913 (2016).
Swarts, D.C., van der Oost, J. & Jinek, M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell 66, 221–233.e224 (2017).
Stella, S., Alcon, P. & Montoya, G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature 546, 559–563 (2017).
Li, B. et al. Engineering CRISPR–Cpf1 crRNAs and mRNAs to maximize genome editing efficiency. Nat. Biomed. Eng. 1, 0066 (2017).
Corey, D.R. Chemical modification: the key to clinical application of RNA interference? J. Clin. Invest. 117, 3615–3622 (2007).
Deleavey, G.F. & Damha, M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 19, 937–954 (2012).
Burnett, J.C. & Rossi, J.J. RNA-based therapeutics: current progress and future prospects. Chem. Biol. 19, 60–71 (2012).
Li, B., Luo, X. & Dong, Y. Effects of chemically modified messenger RNA on protein expression. Bioconjug. Chem. 27, 849–853 (2016).
Dowdy, S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 35, 222–229 (2017).
Labun, K., Montague, T.G., Gagnon, J.A., Thyme, S.B. & Valen, E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–276 (2016).
Kim, H.K. et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods 14, 153–159 (2017).
Bae, S., Park, J. & Kim, J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Cradick, T.J., Qiu, P., Lee, C.M., Fine, E.J. & Bao, G. COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol. Ther. Nucleic Acids 3, e214 (2014).
Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015).
Rahdar, M. et al. Synthetic CRISPR RNA-Cas9–guided genome editing in human cells. Proc. Natl. Acad. Sci. USA 112, E7110–E7117 (2015).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Dang, Y. et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16, 280 (2015).
Kariko, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008).
Anderson, B.R. et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010).
Kormann, M.S. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 (2011).
Zangi, L. et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 31, 898–907 (2013).
Sahin, U., Kariko, K. & Tureci, O. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014).
Andries, O. et al. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344 (2015).
Thess, A. et al. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 23, 1456–1464 (2015).
Uchida, S., Kataoka, K. & Itaka, K. Screening of mRNA chemical modification to maximize protein expression with reduced immunogenicity. Pharmaceutics 7, 137–151 (2015).
Vouillot, L., Thélie, A. & Pollet, N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3: Genes Genomes Genet. 5, 407–415 (2015).
Goldman, D. & Domschke, K. Making sense of deep sequencing. Int. J. Neuropsychopharmacol. 17, 1717–1725 (2014).
Pinello, L. et al. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 34, 695–697 (2016).
Acknowledgements
We acknowledge F. Zhang and his laboratory at the Broad Institute of MIT and Harvard for providing Cpf1 plasmids and technical assistance. This work was supported by the National Institutes of Health through the National Heart, Lung, and Blood Institute (NHLBI; R01HL136652), as well as by the start-up fund from the College of Pharmacy at The Ohio State University.
Author information
Authors and Affiliations
Contributions
B.L., C.Z., and Y.D. developed the protocol.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Representative examples of chemically modified nucleotides.
(a) Linkage (backbone)-modified nucleotides. (b) Ribose-modified nucleotides. (c) Nitrogenous base (cytosine, uracil, adenine, and guanine)-modified nucleotides. Natural nucleotides are shown in blue. The modified groups are shown in red.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 and Supplementary Table 1. (PDF 742 kb)
Rights and permissions
About this article
Cite this article
Li, B., Zeng, C. & Dong, Y. Design and assessment of engineered CRISPR–Cpf1 and its use for genome editing. Nat Protoc 13, 899–914 (2018). https://doi.org/10.1038/nprot.2018.004
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2018.004
This article is cited by
-
Cas13d knockdown of lung protease Ctsl prevents and treats SARS-CoV-2 infection
Nature Chemical Biology (2022)
-
A short overview of CRISPR-Cas technology and its application in viral disease control
Transgenic Research (2021)
-
Heterologous Expression of Lignocellulose-Modifying Enzymes in Microorganisms: Current Status
Molecular Biotechnology (2021)
-
Switching the activity of Cas12a using guide RNA strand displacement circuits
Nature Communications (2019)
-
Synthetic chimeric nucleases function for efficient genome editing
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.