Abstract
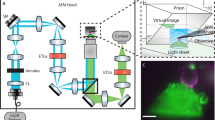
Over the past five years, atomic force microscopy (AFM)-based approaches have evolved into a powerful multiparametric tool set capable of imaging the surfaces of biological samples ranging from single receptors to membranes and tissues. One of these approaches, force–distance curve-based AFM (FD-based AFM), uses a probing tip functionalized with a ligand to image living cells at high-resolution and simultaneously localize and characterize specific ligand–receptor binding events. Analyzing data from FD-based AFM experiments using appropriate probabilistic models allows quantification of the kinetic and thermodynamic parameters that describe the free-energy landscape of the ligand–receptor bond. We have recently developed an FD-based AFM approach to quantify the binding events of single enveloped viruses to surface receptors of living animal cells while simultaneously observing them by fluorescence microscopy. This approach has provided insights into the early stages of the interaction between a virus and a cell. Applied to a model virus, we probed the specific interaction with cells expressing viral cognate receptors and measured the affinity of the interaction. Furthermore, we observed that the virus rapidly established specific multivalent interactions and found that each bond formed in sequence strengthened the attachment of the virus to the cell. Here we describe detailed procedures for probing the specific interactions of viruses with living cells; these procedures cover tip preparation, cell sample preparation, step-by-step FD-based AFM imaging and data analysis. Experienced microscopists should be able to master the entire set of protocols in 1 month.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dimitrov, D.S. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2, 109–122 (2004).
Smith, A.E. & Helenius, A. How viruses enter animal cells. Science 304, 237–242 (2004).
Brandenburg, B. & Zhuang, X. Virus trafficking - learning from single-virus tracking. Nat. Rev. Microbiol. 5, 197–208 (2007).
Alsteens, D. et al. Nanomechanical mapping of first binding steps of a virus to animal cells. Nat. Nanotechnol. 12, 177–183 (2017).
Schnell, M.J., Mebatsion, T. & Conzelmann, K.K. Infectious rabies viruses from cloned cDNA. EMBO J. 13, 4195–4203 (1994).
Ghanem, A., Kern, A. & Conzelmann, K.K. Significantly improved rescue of rabies virus from cDNA plasmids. Eur. J. Cell Biol. 91, 10–16 (2012).
Herrmann, A. & Sieben, C. Single-virus force spectroscopy unravels molecular details of virus infection. Integr. Biol. 7, 620–632 (2015).
Matrosovich, M.N. & Gambaryan, A.S. Solid-phase assays of receptor-binding specificity. Methods Mol. Biol. 865, 71–94 (2012).
Watanabe, T. et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501, 551–555 (2013).
Shi, Y. et al. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342, 243–247 (2013).
Suenaga, E., Mizuno, H. & Penmetcha, K.K Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens. Bioelectron. 32, 195–201 (2012).
Papp, I. et al. Inhibition of influenza virus infection by multivalent sialic-acid-functionalized gold nanoparticles. Small 6, 2900–2906 (2010).
Xiong, X. et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497, 392–396 (2013).
Roingeard, P. Viral detection by electron microscopy: past, present and future. Biol. Cell 100, 491–501 (2008).
Mercer, J. & Helenius, A. Virus entry by macropinocytosis. Nat. Cell Biol. 11, 510–520 (2009).
Ando, T., Uchihashi, T. & Kodera, N. High-speed AFM and applications to biomolecular systems. Annu. Rev. Biophys. 42, 393–414 (2013).
Kienberger, F., Mueller, H., Pastushenko, V. & Hinterdorfer, P. Following single antibody binding to purple membranes in real time. EMBO Rep. 5, 579–583 (2004).
Hinterdorfer, P. & Dufrêne, Y.F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 3, 347–355 (2006).
Neuman, K.C. & Nagy, A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 5, 491–505 (2008).
Sieben, C. et al. Influenza virus binds its host cell using multiple dynamic interactions. Proc. Natl. Acad. Sci. USA 109, 13626–13631 (2012).
Rankl, C. et al. Multiple receptors involved in human rhinovirus attachment to live cells. Proc. Natl. Acad. Sci. USA 105, 17778–17783 (2008).
Chang, M.I., Panorchan, P., Dobrowsky, T.M., Tseng, Y. & Wirtz, D. Single-molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells. J. Virol. 79, 14748–14755 (2005).
Dobrowsky, T.M., Zhou, Y., Sun, S.X., Siliciano, R.F. & Wirtz, D. Monitoring early fusion dynamics of human immunodeficiency virus type 1 at single-molecule resolution. J. Virol. 82, 7022–7033 (2008).
Raab, A. et al. Antibody recognition imaging by force microscopy. Nat. Biotechnol. 17, 901–905 (1999).
Ludwig, M., Dettmann, W. & Gaub, H. Atomic force microscope imaging contrast based on molecular recognition. Biophys. J. 72, 445–448 (1997).
Pfreundschuh, M., Martinez-Martin, D., Mulvihill, E., Wegmann, S. & Muller, D.J. Multiparametric high-resolution imaging of native proteins by force-distance curve–based AFM. Nat. Protoc. 9, 1113–1130 (2014).
Sun, E., He, J. & Zhuang, X. Live cell imaging of viral entry. Curr. Opin. Virol. 3, 34–43 (2013).
Dufrêne, Y.F., Martinez-Martin, D., Medalsy, I., Alsteens, D. & Muller, D.J. Multiparametric imaging of biological systems by force-distance curve-based AFM. Nat. Methods 10, 847–854 (2013).
Sieben, C. & Herrmann, A. Single virus force spectroscopy: the ties that bind. Nat. Nanotechnol. 12, 102–103 (2017).
Engel, A. & Müller, D.J. Observing single biomolecules at work with the atomic force microscope. Nat. Struct. Mol. Biol. 7, 715–718 (2000).
Xiao, J. & Dufrêne, Y.F. Optical and force nanoscopy in microbiology. Nat. Microbiol. 1, 16186 (2016).
Alsteens, D., Trabelsi, H., Soumillion, P. & Dufrêne, Y.F. Multiparametric atomic force microscopy imaging of single bacteriophages extruding from living bacteria. Nat. Commun. 4 (2013).
Puntheeranurak, T., Neundlinger, I., Kinne, R.K.H. & Hinterdorfer, P. Single-molecule recognition force spectroscopy of transmembrane transporters on living cells. Nat. Protoc. 6, 1443–1452 (2011).
Evans, E.A. & Calderwood, D.A. Forces and bond dynamics in cell adhesion. Science 316, 1148–1153 (2007).
Evans, E. & Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541–1555 (1997).
Alsteens, D. et al. High-resolution imaging of chemical and biological sites on living cells using peak force tapping atomic force microscopy. Langmuir 28, 16738–16744 (2012).
Parachoniak, C.A. & Park, M. Dynamics of receptor trafficking in tumorigenicity. Trends Cell Biol. 22, 231–240 (2012).
Reichl, E.M., Effler, J.C. & Robinson, D.N. The stress and strain of cytokinesis. Trends Cell Biol. 15, 200–206 (2005).
Huang, S. & Ingber, D.E. The structural and mechanical complexity of cell-growth control. Nat. Cell Biol. 1, E131–E138 (1999).
Janmey, P.A. The cytoskeleton and cell signaling: component localization and mechanical coupling. Physiol. Rev. 78, 763–781 (1998).
Parsons, J.T., Horwitz, A.R. & Schwartz, M.A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 (2010).
Bao, G. & Suresh, S. Cell and molecular mechanics of biological materials. Nat. Mater. 2, 715–725 (2003).
Matias, V. & Beveridge, T. Cryoelectron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56, 240–251 (2005).
Hell, S.W. Far-field optical nanoscopy. Science 316, 1153–1158 (2007).
Alsteens, D. et al. Atomic force microscopy-based characterization and design of biointerfaces. Nat. Rev. Mat. 2, 17008 (2017).
Dufrene, Y.F. et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 12, 295–307 (2017).
Schillers, H., Medalsy, I., Hu, S., Slade, A.L. & Shaw, J.E. PeakForce Tapping resolves individual microvilli on living cells. J. Mol. Recognit. 29, 95–101 (2016).
Iyer, S., Gaikwad, R., Subba-Rao, V., Woodworth, C. & Sokolov, I. Atomic force microscopy detects differences in the surface brush of normal and cancerous cells. Nat. Nanotechnol. 4, 389–393 (2009).
Krieg, M. et al. Tensile forces govern germ-layer organization in zebrafish. Nat. Cell Biol. 10, 429–436 (2008).
Krieg, M., Dunn, A.R. & Goodman, M.B. Mechanical control of the sense of touch by β-spectrin. Nat. Cell Biol. 16, 224–233 (2014).
Matzke, R., Jacobson, K. & Radmacher, M. Direct, high-resolution measurement of furrow stiffening during division of adherent cells. Nat. Cell Biol. 3, 607–610 (2001).
Stewart, M.P. et al. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature 469, 226–230 (2011).
Hanrahan, J. & Tabcharani, J. Inhibition of an outwardly rectifying anion channel by HEPES and related buffers. J. Membr. Biol. 116, 65–77 (1990).
Lepe-Zuniga, J.L., Zigler, J. & Gery, I. Toxicity of light-exposed Hepes media. J. Immunol. Methods 103, 145 (1987).
Otero, D.H., Wilbekin, F. & Meyer, E.M. Effects of 4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid (AH5183) on rat cortical synaptosome choline uptake, acetylcholine storage and release. Brain Res. 359, 208–214 (1985).
Papp, I. et al. Inhibition of influenza virus activity by multivalent glycoarchitectures with matched sizes. Chem. Bio. Chem. 12, 887–895 (2011).
Watabe-Uchida, M., Zhu, L., Ogawa, S.K., Vamanrao, A. & Uchida, N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron 74, 858–873 (2012).
Barde, I., Salmon, P. & Trono, D. Production and titration of lentiviral vectors. Curr. Protoc. Neurosci. 4, Unit 4 21 (2010).
Wickersham, I.R., Sullivan, H.A. & Seung, H.S. Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. Nat. Protoc. 5, 595–606 (2010).
Wildling, L. et al. Linking of sensor molecules with amino groups to amino-functionalized AFM tips. Bioconjug. Chem. 22, 1239–1248 (2011).
Bell, G.I. Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978).
Collin, D. et al. Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature 437, 231–234 (2005).
Friddle, R.W., Noy, A. & De Yoreo, J.J. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc. Natl. Acad. Sci. USA 109, 13573–13578 (2012).
Sulchek, T., Friddle, R.W. & Noy, A. Strength of multiple parallel biological bonds. Biophys. J. 90, 4686–4691 (2006).
Alsteens, D. et al. Imaging G protein-coupled receptors while quantifying their ligand-binding free-energy landscape. Nat. Methods 12, 845–851 (2015).
Bustamante, C., Marko, J.F., Siggia, E.D. & Smith, S. Entropic elasticity of lambda-phage DNA. Science 265, 1599–1600 (1994).
Narayan, S., Barnard, R.J.O. & Young, J.A.T. Two retroviral entry pathways distinguished by lipid raft association of the viral receptor and differences in viral infectivity. J. Virol. 77, 1977–1983 (2003).
Osakada, F. et al. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron 71, 617–631 (2011).
Gomme, E.A., Faul, E.J., Flomenberg, P., McGettigan, J.P. & Schnell, M.J. Characterization of a single-cycle rabies virus-based vaccine vector. J. Virol. 84, 2820–2831 (2010).
Wickersham, I.R. et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647 (2007).
Butt, H.J. & Jaschke, M. Calculation of thermal noise in atomic-force microscopy. Nanotechnology 6, 1–7 (1995).
Burnham, N.A. et al. Comparison of calibration methods for atomic-force microscopy cantilevers. Nanotechnology 14, 1–6 (2003).
Janovjak, H., Struckmeier, J. & Muller, D.J. Hydrodynamic effects in fast AFM single-molecule force measurements. Eur. Biophys. J. 34, 91–96 (2005).
Alcaraz, J. et al. Correction of microrheological measurements of soft samples with atomic force microscopy for the hydrodynamic drag on the cantilever. Langmuir 18, 716–721 (2002).
Bizzarri, A.R. & Cannistraro, S. The application of atomic force spectroscopy to the study of biological complexes undergoing a biorecognition process. Chem. Soc. Rev. 39, 734–749 (2010).
Hane, F., Attwood, S. & Leonenko, Z. Comparison of three competing dynamic force spectroscopy models to study binding forces of amyloid-β (1–42). Soft Matter 10, 1924–1930 (2014).
Mammen, M., Choi, S.-K. & Whitesides, G.M. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. Engl. 37, 2754–2794 (1998).
Evans, E. & Williams,, P. in Physics of Bio-Molecules and cells (eds H. Flyvbjerg, F. Jülicher, P. Orms, & F. David) 145–204 (ringer, 2002).
Damico, R.L., Crane, J. & Bates, P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc. Natl. Acad. Sci. USA 95, 2580–2585 (1998).
Acknowledgements
We thank our colleagues and collaborators for sharing exciting experiments and discussions. This protocol owes much to previous work from the labs of S. Seung (MIT; ΔGRabies propagation), D. Trono (École polytechnique fédérale de Lausanne; lentivirus production) and H. Gruber (Johannes Kepler University Linz; cantilever functionalization). We thank K. Yonehara (Aarhus University) and members of the B. Roska lab (Friedrich Miescher Institute), particularly J. Jüttner and K. Balint, for their valuable help in modifying the virus production protocols to suit our needs. We thank K. Conzelmann (Ludwig-Maximilans University), S. Finke. (Friedrich-Loeffler Institute) and B. Roska for kindly providing stocks of ΔGRabies. The plasmid pAAV-EF1α-FLEX-TVA-mCherry was a gift from N. Uchida (Harvard University) and the plasmids pRRLSIN.cppt.PGK-GFP.WPRE, pMD2.G and pCMV-dR8.74 were gifts from D. Trono. We thank M. Mohr (ETH Zurich) for assistance in sub-cloning the pRRLSIN.cppt.EF1α plasmid. This work was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the 'MOVE-IN Louvain' incoming post-doc fellowship programme, the Swiss National Science Foundation (SNF; grant no. 310030B_160225) and NCCR Molecular Systems Engineering. D.A. is a research associate at the FRS-FNRS.
Author information
Authors and Affiliations
Contributions
R.N., M.D., D.J.M. and D.A. designed and performed the experiments. R.N., M.D., M.K., A.C.D., P.R.L., D.J.M. and D.A. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
D.J.M. and D.A. have applied for a patent for the chamber enabling AFM and optical microscopy under cell-culture conditions (EP15002176.4). The other authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Newton, R., Delguste, M., Koehler, M. et al. Combining confocal and atomic force microscopy to quantify single-virus binding to mammalian cell surfaces. Nat Protoc 12, 2275–2292 (2017). https://doi.org/10.1038/nprot.2017.112
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.112
This article is cited by
-
Paired immunoglobulin-like receptor B is an entry receptor for mammalian orthoreovirus
Nature Communications (2023)
-
Multivalent 9-O-Acetylated-sialic acid glycoclusters as potent inhibitors for SARS-CoV-2 infection
Nature Communications (2022)
-
Force spectroscopy of single cells using atomic force microscopy
Nature Reviews Methods Primers (2021)
-
Reovirus directly engages integrin to recruit clathrin for entry into host cells
Nature Communications (2021)
-
Atomic force microscopy for revealing micro/nanoscale mechanics in tumor metastasis: from single cells to microenvironmental cues
Acta Pharmacologica Sinica (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.