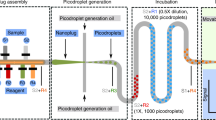
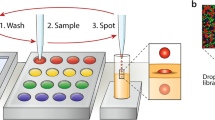
Abstract
Biochemical systems in which multiple components take part in a given reaction are of increasing interest. Because the interactions between these different components are complex and difficult to predict from basic reaction kinetics, it is important to test for the effect of variations in the concentration for each reagent in a combinatorial manner. For example, in PCR, an increase in the concentration of primers initially increases template amplification, but large amounts of primers result in primer–dimer by-products that inhibit the amplification of the template. Manual titration of biochemical mixtures rapidly becomes costly and laborious, forcing scientists to settle for suboptimal concentrations. Here we present a droplet-based microfluidics platform for mapping of the concentration space of up to three reaction components followed by detection with a fluorescent readout. The concentration of each reaction component is read through its internal standard (barcode), which is fluorescent but chemically orthogonal. We describe in detail the workflow, which comprises the following: (i) production of the microfluidics chips, (ii) preparation of the biochemical mixes, (iii) their mixing and compartmentalization into water-in-oil emulsion droplets via microfluidics, (iv) incubation and imaging of the fluorescent barcode and reporter signals by fluorescence microscopy and (v) image processing and data analysis. We also provide recommendations for choosing the appropriate fluorescent markers, programming the pressure profiles and analyzing the generated data. Overall, this platform allows a researcher with a few weeks of training to acquire ∼10,000 data points (in a 1D, 2D or 3D concentration space) over the course of a day from as little as 100–1,000 μl of reaction mix.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Burlingham, B.T. & Widlanski, T.S. An intuitive look at the relationship of Ki and IC50: a more general use for the Dixon plot. J. Chem. Educ. 80, 214–218 (2003).
Zimmer, A., Katzir, I., Dekel, E., Mayo, A.E. & Alon, U. Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc. Natl. Acad. Sci. USA 113, 10442–10447 (2016).
Elnifro, E.M., Ashshi, A.M., Cooper, R.J. & Klapper, P.E. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 13, 559–570 (2000).
SantaLucia, J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95, 1460–1465 (1998).
Zadeh, J.N. et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011).
Bustin, S.A. Why the need for qPCR publication guidelines? The case for MIQE. Methods 50, 217–226 (2010).
Klingenberg, M. The original micropipette. Scientist 20, 92–92 (2006).
Unger, M.A., Chou, H.P., Thorsen, T., Scherer, A. & Quake, S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116 (2000).
Thorsen, T., Maerkl, S.J. & Quake, S.R. Microfluidic large-scale integration. Science 298, 580–584 (2002).
Sackmann, E.K., Fulton, A.L. & Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 507, 181–189 (2014).
Vogelstein, B. & Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 96, 9236–9241 (1999).
Rondelez, Y. et al. Microfabricated arrays of femtoliter chambers allow single molecule enzymology. Nat. Biotechnol. 23, 361–365 (2005).
Ottesen, E.A., Hong, J.W., Quake, S.R. & Leadbetter, J.R. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science 314, 1464–1467 (2006).
Pekin, D. et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip 11, 2156–2166 (2011).
Kim, S.H. et al. Large-scale femtoliter droplet array for digital counting of single biomolecules. Lab Chip 12, 4986–4991 (2012).
Witters, D. et al. Digital biology and chemistry. Lab Chip 14, 3225–3232 (2014).
Petersen, K.E. Silicon as a mechanical material. Proc. IEEE 70, 420–457 (1982).
Duffy, D.C., McDonald, J.C., Schueller, O.J.A. & Whitesides, G.M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998).
Teh, S.-Y., Lin, R., Hung, L.-H. & Lee, A.P. Droplet microfluidics. Lab Chip 8, 198–220 (2008).
Leman, M., Abouakil, F., Griffiths, A.D. & Tabeling, P. Droplet-based microfluidics at the femtolitre scale. Lab Chip 15, 753–765 (2015).
Shim, J.-U. et al. Ultrarapid generation of femtoliter microfluidic droplets for single-molecule-counting immunoassays. ACS Nano 7, 5955–5964 (2013).
Bringer, M.R., Gerdts, C.J., Song, H., Tice, J.D. & Ismagilov, R.F. Microfluidic systems for chemical kinetics that rely on chaotic mixing in droplets. Philos. Trans. A Math. Phys. Eng. Sci. 362, 1087–1104 (2004).
Agresti, J.J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 107, 4004–4009 (2010).
Abate, A.R., Hung, T., Mary, P., Agresti, J.J. & Weitz, D.A. High-throughput injection with microfluidics using picoinjectors. Proc. Natl. Acad. Sci. USA 107, 19163–19166 (2010).
Guo, M.T., Rotem, A., Heyman, J.A. & Weitz, D.A. Droplet microfluidics for high-throughput biological assays. Lab Chip 12, 2146–2155 (2012).
Hasatani, K. et al. High-throughput and long-term observation of compartmentalized biochemical oscillators. Chem. Commun. 49, 8090–8092 (2013).
Bai, Y. et al. A double droplet trap system for studying mass transport across a droplet-droplet interface. Lab Chip 10, 1281–1285 (2010).
Garstecki, P., Fuerstman, M.J., Stone, H.A. & Whitesides, G.M. Formation of droplets and bubbles in a microfluidic T-junction—scaling and mechanism of break-up. Lab Chip 6, 437–446 (2006).
Kaneda, S. et al. Pneumatic handling of droplets on-demand on a microfluidic device for seamless processing of reaction and electrophoretic separation. Electrophoresis 31, 3719–3726 (2010).
Kim, J.H. et al. Droplet microfluidics for producing functional microparticles. Langmuir 30, 1473–1488 (2014).
Caen, O. et al. Digital PCR compartmentalization II. Contribution for the quantitative detection of circulating tumor DNA. Med. Sci. 31, 180–186 (2015).
Yamashita, H. et al. Generation of monodisperse cell-sized microdroplets using a centrifuge-based axisymmetric co-flowing microfluidic device. J. Biosci. Bioeng. 119, 492–495 (2015).
Anna, S.L. Droplets and bubbles in microfluidic devices. Annu. Rev. Fluid Mech. 48, 285–309 (2016).
Baroud, C.N., Gallaire, F. & Dangla, R. Dynamics of microfluidic droplets. Lab Chip 10, 2032–2045 (2010).
Nakano, M. et al. Single-molecule PCR using water-in-oil emulsion. J. Biotechnol. 102, 117–124 (2003).
Williams, R. et al. Amplification of complex gene libraries by emulsion PCR. Nat. Methods 3, 545–550 (2006).
Weitz, M. et al. Diversity in the dynamical behaviour of a compartmentalized programmable biochemical oscillator. Nat. Chem. 6, 295–302 (2014).
Thorsen, T., Roberts, R.W., Arnold, F.H. & Quake, S.R. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 86, 4163–4166 (2001).
Song, H., Chen, D.L. & Ismagilov, R.F. Reactions in droplets in microfluidic channels. Angew. Chem. Int. Ed. 45, 7336–7356 (2006).
Ward, T., Faivre, M., Abkarian, M. & Stone, H.A. Microfluidic flow focusing: drop size and scaling in pressure versus flow-rate-driven pumping. Electrophoresis 26, 3716–3724 (2005).
Taly, V., Kelly, B.T. & Griffiths, A.D. Droplets as microreactors for high-throughput biology. ChemBioChem 8, 263–272 (2007).
Mazutis, L. et al. Multi-step microfluidic droplet processing: kinetic analysis of an in vitro translated enzyme. Lab Chip 9, 2902–2908 (2009).
Pompano, R.R., Liu, W.S., Du, W.B. & Ismagilov, R.F. Microfluidics using spatially defined arrays of droplets in one, two, and three dimensions. Annu. Rev. Anal. Chem. 4, 59–81 (2011).
Taly, V. et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 59, 1722–1731 (2013).
Lim, J. et al. Parallelized ultra-high throughput microfluidic emulsifier for multiplex kinetic assays. Biomicrofluidics 9 (2015).
Sugiura, H. et al. Pulse-density modulation control of chemical oscillation far from equilibrium in a droplet open-reactor system. Nat. Commun. 7, 10212 (2016).
Matsumura, S. et al. Transient compartmentalization of RNA replicators prevents extinction due to parasites. Science 354, 1293–1296 (2016).
Eisenstein, M. Startups use short-read data to expand long-read sequencing market. Nat. Biotechnol. 33, 433–435 (2015).
Baker, M. Digital PCR hits its stride. Nat. Methods 9, 541–544 (2012).
Miller, O.J. et al. High-resolution dose-response screening using droplet-based microfluidics. Proc. Natl. Acad. Sci. USA 109, 378–383 (2012).
Zheng, B., Roach, L.S. & Ismagilov, R.F. Screening of protein crystallization conditions on a microfluidic chip using nanoliter-size droplets. J. Am. Chem. Soc. 125, 11170–11171 (2003).
Genot, A.J. et al. High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nat. Chem. 8, 760–767 (2016).
Notomi, T. et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, e63 (2000).
Van Ness, J., Van Ness, L.K. & Galas, D.J. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl. Acad. Sci. USA 100, 4504–4509 (2003).
Johnson, K.A. & Goody, R.S. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry 50, 8264–8269 (2011).
Borch, J. & Roepstorff, P. Screening for enzyme inhibitors by surface plasmon resonance combined with mass spectrometry. Anal. Chem. 76, 5243–5248 (2004).
Fredriksson, S. et al. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 (2002).
Shults, M.D., Janes, K.A., Lauffenburger, D.A. & Imperiali, B. A multiplexed homogeneous fluorescence-based assay for protein kinase activity in cell lysates. Nat. Methods 2, 277–283 (2005).
Geladopoulos, T.P., Sotiroudis, T.G. & Evangelopoulos, A.E. A malachite green colorimetric assay for protein phosphatase-activity. Anal. Biochem. 192, 112–116 (1991).
Pelletier, J., Bellot, G., Pouysségur, J. & Mazure, N.M. Biochemical titration of glycogen in vitro. J. Vis. Exp. 2013, e50465 (2013).
Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Shendure, J. & Ji, H.L. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008).
Gielen, F. et al. Ultrahigh-throughput-directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proc. Natl. Acad. Sci. USA 113, E7383–E7389 (2016).
Gruner, P. et al. Stabilisers for water-in-fluorinated-oil dispersions: key properties for microfluidic applications. Curr. Opin. Colloid Interface Sci. 20, 183–191 (2015).
Gu, S.Q. et al. Droplet-based microfluidics for dose-response assay of enzyme inhibitors by electrochemical method. Anal. Chim. Acta 796, 68–74 (2013).
Gerver, R.E. et al. Programmable microfluidic synthesis of spectrally encoded microspheres. Lab Chip 12, 4716–4723 (2012).
Cai, L.F., Zhu, Y., Du, G.S. & Fang, Q. Droplet-based microfluidic flow injection system with large-scale concentration gradient by a single nanoliter-scale injection for enzyme inhibition assay. Anal. Chem. 84, 446–452 (2012).
Theberge, A.B. et al. Microfluidic platform for combinatorial synthesis in picolitre droplets. Lab Chip 12, 1320–1326 (2012).
Niu, X., Gielen, F., Edel, J.B. & deMello, A.J. A microdroplet dilutor for high-throughput screening. Nat. Chem. 3, 437–442 (2011).
Sun, M., Bithi, S.S. & Vanapalli, S.A. Microfluidic static droplet arrays with tuneable gradients in material composition. Lab Chip 11, 3949–3952 (2011).
Bui, M.P.N. et al. Enzyme kinetic measurements using a droplet-based microfluidic system with a concentration gradient. Anal. Chem. 83, 1603–1608 (2011).
Du, W.B., Sun, M., Gu, S.Q., Zhu, Y. & Fang, Q. Automated microfluidic screening assay platform based on drop lab. Anal. Chem. 82, 9941–9947 (2010).
Du, W.B., Li, L., Nichols, K.P. & Ismagilov, R.F. SlipChip. Lab Chip 9, 2286–2292 (2009).
Damean, N., Olguin, L.F., Hollfelder, F., Abell, C. & Huck, W.T.S. Simultaneous measurement of reactions in microdroplets filled by concentration gradients. Lab Chip 9, 1707–1713 (2009).
Lorenz, R.M. et al. Simultaneous generation of multiple aqueous droplets in a microfluidic device. Anal. Chim. Acta 630, 124–130 (2008).
Song, H. & Ismagilov, R.F. Millisecond kinetics on a microfluidic chip using nanoliters of reagents. J. Am. Chem. Soc. 125, 14613–14619 (2003).
Tabeling, P. Introduction to Microfluidics (Oxford University Press, 2005).
Kijani, S. et al. Filter-dense multicolor microscopy. PLoS One 10 (2015).
Paik, P., Pamula, V.K. & Fair, R.B. Rapid droplet mixers for digital microfluidic systems. Lab Chip 3, 253–259 (2003).
Niu, X., Gulati, S., Edel, J.B. & deMello, A.J. Pillar-induced droplet merging in microfluidic circuits. Lab Chip 8, 1837–1841 (2008).
Courtois, F. et al. Controlling the retention of small molecules in emulsion microdroplets for use in cell-based assays. Anal. Chem. 81, 3008–3016 (2009).
Skhiri, Y. et al. Dynamics of molecular transport by surfactants in emulsions. Soft Matter 8, 10618–10627 (2012).
Gruner, P. et al. Controlling molecular transport in minimal emulsions. Nat. Commun. 7, 10392 (2016).
Hofmann, T.W., Anselmann, S.H., Janiesch, J.W., Rademacher, A. & Bohm, C.H.J. Applying microdroplets as sensors for label-free detection of chemical reactions. Lab Chip 12, 916–922 (2012).
Li, X.B., Li, F.C., Kinoshita, H., Oishi, M. & Oshima, M. Dynamics of viscoelastic fluid droplet under very low interfacial tension in a serpentine T-junction microchannel. Microfluid. Nanofluid. 18, 1007–1021 (2015).
Qin, D., Xia, Y.N. & Whitesides, G.M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 5, 491–502 (2010).
Mazutis, L. et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 8, 870–891 (2013).
Kaneda, S. et al. Modification of the glass surface property in PDMS-glass hybrid microfluidic devices. Anal. Sci. 28, 39–44 (2012).
Baret, J.C. Surfactants in droplet-based microfluidics. Lab Chip 12, 422–433 (2012).
Holtze, C. et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 8, 1632–1639 (2008).
Zhong, Q. et al. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab Chip 11, 2167–2174 (2011).
Baret, J.C. et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip 9, 1850–1858 (2009).
Schmitz, C.H.J., Rowat, A.C., Koster, S. & Weitz, D.A. Dropspots: a picoliter array in a microfluidic device. Lab Chip 9, 44–49 (2009).
Heid, C.A., Stevens, J., Livak, K.J. & Williams, P.M. Real time quantitative PCR. Genome Res. 6, 986–994 (1996).
Schmidt, M. & Lipson, H. Distilling free-form natural laws from experimental data. Science 324, 81–85 (2009).
Cully, A., Clune, J., Tarapore, D. & Mouret, J.B. Robots that can adapt like animals. Nature 521, 503–507 (2015).
Deb, K., Pratap, A., Agarwal, S. & Meyarivan, T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 6, 182–197 (2002).
Kask, P., Palo, K., Hinnah, C. & Pommerencke, T. Flat field correction for high-throughput imaging of fluorescent samples. J. Microsc. 263, 328–340 (2016).
Tanaka, H. et al. Hands-off preparation of monodisperse emulsion droplets using a poly(dimethylsiloxane) microfluidic chip for droplet digital PCR. Anal. Chem. 87, 4134–4143 (2015).
Pandit, K.R., Rueger, P.E., Calabrese, R.V., Raghavan, S.R. & White, I.M. Assessment of surfactants for efficient droplet PCR in mineral oil using the pendant drop technique. Colloids Surf. B Biointerfaces 126, 489–495 (2015).
Acknowledgements
We thank S. Nishikawa and K. Montagne for kindly proofreading the manuscript. Y.R. acknowledges support from the Japan Society for the Promotion of Science (JSPS) through a Grant-in-Aid for Scientific Research on Innovative Areas 'Synthetic Biology for Comprehension of Biomolecular Networks' (project number 23119001). A.J.G. acknowledges support from the ANR (ANR-13-PDOC-0001) and the JSPS (postdoctoral fellowship). J.-F.B. was supported by a PhD fellowship from Region Alsace. N.B. acknowledges support from the PHC Sakura program (project number 34171WG). A.B. and N.A.-K. were supported by the ELSI Origins Network (EON), which is supported by a grant from the John Templeton Foundation. The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation.
Author information
Authors and Affiliations
Contributions
All authors contributed to the development of the method. A.B., S.O., R.S., E.H. and A.J.G. performed the experiments. N.B., N.A.-K., Y.R. and A.J.G. analyzed the data. J.-F.B. and V.T. synthesized the surfactant. T.F. and V.T. provided support with the microfluidic platform. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data 1
An SU-8 master mold of the chip array (ZIP file). (ZIP 507 kb)
Supplementary Data 2
Pressure scripts (ZIP file). (ZIP 70 kb)
Supplementary Data 3
MATLAB script (TXT file). (TXT 4 kb)
Rights and permissions
About this article
Cite this article
Baccouche, A., Okumura, S., Sieskind, R. et al. Massively parallel and multiparameter titration of biochemical assays with droplet microfluidics. Nat Protoc 12, 1912–1932 (2017). https://doi.org/10.1038/nprot.2017.092
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.092
This article is cited by
-
Tunable encapsulation of sessile droplets with solid and liquid shells
Nature Communications (2023)
-
Nonlinear decision-making with enzymatic neural networks
Nature (2022)
-
Microfluidic droplet generation based on non-embedded co-flow-focusing using 3D printed nozzle
Scientific Reports (2020)
-
Designing Dynamical Molecular Systems with the PEN Toolbox
New Generation Computing (2020)
-
Microfluidic device for real-time formulation of reagents and their subsequent encapsulation into double emulsions
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.