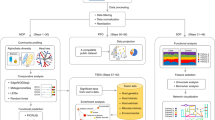
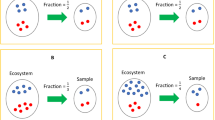
Abstract
Over the last decade, technical advances in nucleic acid sequencing and mass spectrometry have enabled faster and more informative metagenomic, metatranscriptomic, metaproteomic and metabolomic measurements. Here we review key improvements in multi-omic environmental and human microbiome analyses, and discuss developments required to address current measurement shortcomings.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Locey, K.J. & Lennon, J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA 113, 5970–5975 (2016).
Jansson, J.K. Towards “Tera-Terra”: Terabase sequencing of terrestrial metagenomes. eScholarship http://escholarship.org/uc/item/04p1x29k##2011.
Lamendella, R., VerBerkmoes, N. & Jansson, J.K. 'Omics' of the mammalian gut—new insights into function. Curr. Opin. Biotechnol. 23, 491–500 (2012).
Gilbert, J.A., Jansson, J.K. & Knight, R. The Earth Microbiome project: successes and aspirations. BMC Biol. 12, 69 (2014).
Alivisatos, A.P. et al. Microbiome. A unified initiative to harness Earth's microbiomes. Science 350, 507–508 (2015).
Lozupone, C.A., Stombaugh, J.I., Gordon, J.I., Jansson, J.K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Heather, J.M. & Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 107, 1–8 (2016).
Sanger, F., Nicklen, S. & Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467 (1977).
Fleischmann, R.D. et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 (1995).
Blattner, F.R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997).
Loman, N.J. et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 30, 434–439 (2012).
Ronaghi, M., Uhlén, M. & Nyrén, P. A sequencing method based on real-time pyrophosphate. Science 281, 363, 365 (1998).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Bentley, D.R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Voskoboynik, A. et al. The genome sequence of the colonial chordate, Botryllus schlosseri. eLife 2, e00569 (2013).
White, R.A. III et al. Moleculo long-read sequencing facilitates assembly and genomic binning from complex soil metagenomes. mSystems 1, e00045–16 (2016).
Sharon, I. et al. Accurate, multi-kb reads resolve complex populations and detect rare microorganisms. Genome Res. 25, 534–543 (2015).
Kuleshov, V. et al. Synthetic long-read sequencing reveals intraspecies diversity in the human microbiome. Nat. Biotechnol. 34, 64–69 (2016).
Levene, M.J. et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299, 682–686 (2003).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Murray, I.A. et al. The methylomes of six bacteria. Nucleic Acids Res. 40, 11450–11462 (2012).
Laver, T. et al. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 3, 1–8 (2015).
Venter, J.C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004).
Willing, B.P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854 e1 (2010).
Mason, O.U. et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 6, 1715–1727 (2012).
Mason, O.U. et al. Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J. 8, 1464–1475 (2014).
Mackelprang, R. et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480, 368–371 (2011).
Hultman, J. et al. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature 521, 208–212 (2015).
Goltsman, D.S.A., Comolli, L.R., Thomas, B.C. & Banfield, J.F. Community transcriptomics reveals unexpected high microbial diversity in acidophilic biofilm communities. ISME J. 9, 1014–1023 (2015).
Zoetendal, E.G. et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 6, 1415–1426 (2012).
Shi, Y., Tyson, G.W., Eppley, J.M. & DeLong, E.F. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 5, 999–1013 (2011).
Frias-Lopez, J. et al. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. USA 105, 3805–3810 (2008).
Shi, Y., Tyson, G.W. & DeLong, E.F. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459, 266–269 (2009).
Gilbert, J.A. et al. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3, e3042 (2008).
Gilbert, J.A. et al. Metagenomes and metatranscriptomes from the L4 long-term coastal monitoring station in the Western English Channel. Stand. Genomic Sci. 3, 183–193 (2010).
Yamashita, M. & Fenn, J.B. Negative ion source production with electrospray ion source. J. Phys. Chem. 88, 4671–4675 (1984).
Hu, Q., Noll, R.J., Li, H., Makarov, A., Hardman, M. & Graham Cooks, R. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40, 430–443 (2005).
Kelly, R.T., Tolmachev, A.V., Page, J.S., Tang, K. & Smith, R.D. The ion funnel: theory, implementations, and applications. Mass Spectrom. Rev. 29, 294–312 (2010).
Baker, E.S. et al. Advancing the high throughput identification of liver fibrosis protein signatures using multiplexed ion mobility spectrometry. Mol. Cell. Proteomics 13, 1119–1127 (2014).
Baker, E.S. et al. Enhancing bottom-up and top-down proteomic measurements with ion mobility separations. Proteomics 15, 2766–2776 (2015).
Ram, R.J. et al. Community proteomics of a natural microbial biofilm. Science 308, 1915–1920 (2005).
Aylward, F.O. et al. Leucoagaricus gongylophorus produces diverse enzymes for the degradation of recalcitrant plant polymers in leaf-cutter ant fungus gardens. Appl. Environ. Microbiol. 79, 3770–3778 (2013).
Aylward, F.O. et al. Enrichment and broad representation of plant biomass-degrading enzymes in the specialized hyphal swellings of Leucoagaricus gongylophorus, the fungal symbiont of leaf-cutter ants. PLoS One 10, e0134752 (2015).
Huang, E.L. et al. The fungus gardens of leaf-cutter ants undergo a distinct physiological transition during biomass degradation. Environ. Microbiol. Rep. 6, 389–395 (2014).
Burnum, K.E. et al. Proteome insights into the symbiotic relationship between a captive colony of Nasutitermes corniger and its hindgut microbiome. ISME J. 5, 161–164 (2011).
Verberkmoes, N.C. et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 3, 179–189 (2009).
Handley, K.M. et al. Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J. 7, 800–816 (2013).
Wilmes, P., Heintz-Bus,. & Bond, P.L. A decade of metaproteomics: where we stand and what the future holds. Proteomics 15, 3409–3417 (2015).
Jansson, J.K. & Baker, E.S. A multi-omic future for microbiome studies. Nat. Microbiol. 1, 16049 (2016).
Acknowledgements
We thank N. Johnson and C. Brislawn for their assistance in preparing the figures. This research was supported by the Pan-omics Program that is funded by the US Department of Energy's Office of Biological and Environmental Research (Genomic Science Program) and the Microbiomes in Transition (MinT) Laboratory Directed Research and Development Initiative at the Pacific Northwest National Laboratory. Pacific Northwest National Laboratory is a multi-program national laboratory operated by Battelle for the Department of Energy under contract DE-AC06-76RL01830.
Author information
Authors and Affiliations
Contributions
R.A.W., S.J.C., R.J.M., E.S.B. and J.K.J. all contributed to this work and commented on the manuscript at all stages.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
White, R., Callister, S., Moore, R. et al. The past, present and future of microbiome analyses. Nat Protoc 11, 2049–2053 (2016). https://doi.org/10.1038/nprot.2016.148
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.148
This article is cited by
-
Effects of environmental factors on release amount of heavy metal and structure of microbial community in sediments
International Journal of Environmental Science and Technology (2022)
-
Revealing taxon-specific heavy metal-resistance mechanisms in denitrifying phosphorus removal sludge using genome-centric metaproteomics
Microbiome (2021)
-
Recent progress in the application of omics technologies in the study of bio-mining microorganisms from extreme environments
Microbial Cell Factories (2021)
-
Microbial iron and carbon metabolism as revealed by taxonomy-specific functional diversity in the Southern Ocean
The ISME Journal (2021)
-
Sequencing barcode construction and identification methods based on block error-correction codes
Science China Life Sciences (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.