Abstract
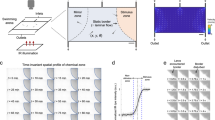
Monitoring neuronal responses to defined sensory stimuli is a powerful and widely used approach for understanding sensory coding in the nervous system. However, providing precise, stereotypic and reproducible cues while concomitantly recording neuronal activity remains technically challenging. Here we describe the fabrication and use of a microfluidics system that allows precise temporally restricted stimulation of Drosophila chemosensory neurons with an array of different chemical cues. The system can easily be combined with genetically encoded calcium sensors, and it can measure neuronal activity at single-cell resolution in larval sense organs and in the proboscis or leg of the adult fly. We describe the design of the master mold, the production of the microfluidic chip and live imaging using the calcium sensor GCaMP, expressed in distinct types of Drosophila chemosensory neurons. Fabrication of the master mold and microfluidic chips requires basic skills in photolithography and takes ∼2 weeks; the same devices can be used repeatedly over several months. Flies can be prepared for measurements in minutes and imaged for up to 1 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
de Bruyne, M., Foster, K. & Carlson, J.R. Odor coding in the Drosophila antenna. Neuron 30, 537–552 (2001).
Benton, R., Sachse, S., Michnick, S.W. & Vosshall, L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4, e20 (2006).
Lee, Y., Moon, S.J. & Montell, C. Multiple gustatory receptors required for the caffeine response in Drosophila. Proc. Natl. Acad. Sci. USA 106, 4495–4500 (2009).
Weiss, L.A., Dahanukar, A., Kwon, J.Y., Banerjee, D. & Carlson, J.R. The molecular and cellular basis of bitter taste in Drosophila. Neuron 69, 258–272 (2011).
Root, C.M., Ko, K.I., Jafari, A. & Wang, J.W. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 145, 133–144 (2011).
Mishra, D. et al. The molecular basis of sugar sensing in Drosophila larvae. Curr. Biol. 23, 1466–1471 (2013).
Zhang, Y.V., Ni, J. & Montell, C. The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338 (2013).
Clyne, P., Grant, A., O'Connell, R. & Carlson, J.R. Odorant response of individual sensilla on the Drosophila antenna. Inver. Neurosci. 3, 127–135 (1997).
Hiroi, M., Marion-Poll, F. & Tanimura, T. Differentiated response to sugars among labellar chemosensilla in Drosophila. Zoolog. Sci. 19, 1009–1018 (2002).
Hallem, E.A., Ho, M.G. & Carlson, J.R. The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979 (2004).
Hallem, E.A. & Carlson, J.R. Coding of odors by a receptor repertoire. Cell 125, 143–160 (2006).
Dahanukar, A., Lei, Y.T., Kwon, J.Y. & Carlson, J.R. Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516 (2007).
Cameron, P., Hiroi, M., Ngai, J. & Scott, K. The molecular basis for water taste in Drosophila. Nature 465, 91–95 (2010).
Vosshall, L.B. & Stocker, R.F. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 30, 505–533 (2007).
Oppliger, F.Y., Guerin, P.M. & Vlimant, M. Neurophysiological and behavioural evidence for an olfactory function for the dorsal organ and a gustatory one for the terminal organ in Drosophila melanogaster larvae. J. Insect Physiol. 46, 135–144 (2000).
Chalasani, S.H. et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 450, 63–70 (2007).
Chronis, N., Zimmer, M. & Bargmann, C.I. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods 4, 727–731 (2007).
Ghannad-Rezaie, M., Wang, X., Mishra, B., Collins, C. & Chronis, N. Microfluidic chips for in vivo imaging of cellular responses to neural injury in Drosophila larvae. PloS One 7, e29869 (2012).
van Giesen, L. et al. Multimodal stimulus coding by a gustatory sensory neuron in Drosophila larvae. Nat. Commun. 7, 10687 (2016).
Liman, E.R., Zhang, Y.V. & Montell, C. Peripheral coding of taste. Neuron 81, 984–1000 (2014).
Choi, J. et al. A pair of pharyngeal gustatory receptor neurons regulates caffeine-dependent ingestion in Drosophila larvae. Front. Cell. Neurosci. 10, 181 (2016).
Brand, A.H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
Chen, T.W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Reiff, D.F. et al. In vivo performance of genetically encoded indicators of neural activity in flies. J. Neurosci. 25, 4766–4778 (2005).
Jayaraman, V. & Laurent, G. Evaluating a genetically encoded optical sensor of neural activity using electrophysiology in intact adult fruit flies. Front. Neural Circuits 1, 3 (2007).
Cao, G. et al. Genetically targeted optical electrophysiology in intact neural circuits. Cell 154, 904–913 (2013).
Gong, Y. et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–1366 (2015).
Tian, L. et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods 6, 875–881 (2009).
Fiala, A. et al. Genetically expressed cameleon in Drosophila melanogaster is used to visualize olfactory information in projection neurons. Curr. Biol. 12, 1877–1884 (2002).
Seelig, J.D. et al. Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat. Methods 7, 535–540 (2010).
Asahina, K., Louis, M., Piccinotti, S. & Vosshall, L.B. A circuit supporting concentration-invariant odor perception in Drosophila. J. Biol. 8, 9 (2009).
Klein, M. et al. Sensory determinants of behavioral dynamics in Drosophila thermotaxis. Proc. Natl. Acad. Sci. USA 112, E220–E229 (2015).
Ni, L. et al. The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. eLife 5, e13254 (2016).
Miyamoto, T., Chen, Y., Slone, J. & Amrein, H. Identification of a Drosophila glucose receptor using Ca2+ imaging of single chemosensory neurons. PloS One 8, e56304 (2013).
Lin, Q. in Physical Properties of Polymers Handbook (ed. J.E. Mark) (2006).
Akerboom, J. et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 32, 13819–13840 (2012).
Fishilevich, E. et al. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr. Biol. 15, 2086–2096 (2005).
Pfeiffer, B.D. et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA 105, 9715–9720 (2008).
Acknowledgements
We thank the Bloomington Drosophila Stock Center for reagents. We thank R. Benton, P. Renaud, L. Pethö, K. Suter and J. Dorsaz for help with the experiments, and T. Graham and B. Egger for helpful discussion of the manuscript. This work was supported by grants from the Swiss National Science Foundation (CRSII3_136307 and 31003A_149499) and the European Research Council (ERC-2012-StG 309832-PhotoNaviNet) to S.G.S. We further thank our colleagues in the Sprecher laboratory for fruitful discussions of the manuscript.
Author information
Authors and Affiliations
Contributions
L.v.G. and G.L.N.-M. performed the experiments. L.v.G. and S.G.S. developed the protocol. L.v.G., G.L.N.-M., J.Y.K. and S.G.S. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Correction of artifacts in the graph.
(a) Representative fluorescent trace before and after movement artifact correction. The fluorescence intensity value before the moving frame was duplicated to mask the artifact. (b) In case of excessive movement the same procedure cannot be applied, and it is recommended to discard the recording.
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 Correction of artifacts in the graph. (a,b) Representative fluorescence trace before and after movement artifact correction. (a) The fluorescence intensity value before the moving frame was duplicated to mask the artifact. (b) In case of excessive movement, the same procedure cannot be applied, and it is recommended that the recording be discarded. (PDF 290 kb)
Supplementary Data
Design of the photomask used for the microfluidic chip fabrication (.cif file). (ZIP 1032 kb)
Rights and permissions
About this article
Cite this article
van Giesen, L., Neagu-Maier, G., Kwon, J. et al. A microfluidics-based method for measuring neuronal activity in Drosophila chemosensory neurons. Nat Protoc 11, 2389–2400 (2016). https://doi.org/10.1038/nprot.2016.144
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.144
This article is cited by
-
An optofluidic platform for interrogating chemosensory behavior and brainwide neural representation in larval zebrafish
Nature Communications (2023)
-
Neuroscience Research using Small Animals on a Chip: From Nematodes to Zebrafish Larvae
BioChip Journal (2021)
-
Tools to reverse-engineer multicellular systems: case studies using the fruit fly
Journal of Biological Engineering (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.