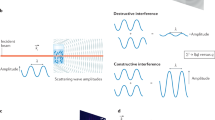
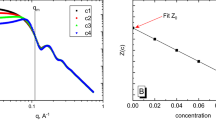
Abstract
Small-angle X-ray scattering (SAXS) and small-angle neutron scattering (SANS) are techniques used to extract structural parameters and determine the overall structures and shapes of biological macromolecules, complexes and assemblies in solution. The scattering intensities measured from a sample contain contributions from all atoms within the illuminated sample volume, including the solvent and buffer components, as well as the macromolecules of interest. To obtain structural information, it is essential to prepare an exactly matched solvent blank so that background scattering contributions can be accurately subtracted from the sample scattering to obtain the net scattering from the macromolecules in the sample. In addition, sample heterogeneity caused by contaminants, aggregates, mismatched solvents, radiation damage or other factors can severely influence and complicate data analysis, so it is essential that the samples be pure and monodisperse for the duration of the experiment. This protocol outlines the basic physics of SAXS and SANS, and it reveals how the underlying conceptual principles of the techniques ultimately 'translate' into practical laboratory guidance for the production of samples of sufficiently high quality for scattering experiments. The procedure describes how to prepare and characterize protein and nucleic acid samples for both SAXS and SANS using gel electrophoresis, size-exclusion chromatography (SEC) and light scattering. Also included are procedures that are specific to X-rays (in-line SEC–SAXS) and neutrons, specifically preparing samples for contrast matching or variation experiments and deuterium labeling of proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout














Similar content being viewed by others
Accession codes
References
Graewert, M.A. & Svergun, D.I. Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS). Curr. Opin. Struct. Biol. 23, 748–754 (2013).
Hura, G.L. et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Meth. 6, 606–612 (2009).
Skou, S., Gillilan, R.E. & Ando, N. Synchrotron-based small-angle X-ray scattering of proteins in solution. Nat. Protoc. 9, 1727–1739 (2014).
Petoukhov, M.V. et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. App. Crystallogr. 45, 342–350 (2012).
Petoukhov, M.V., Konarev, P.V., Kikhney, A.G. & Svergun, D.I. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40, s223–s228 (2007).
Franke, D., Kikhney, A.G. & Svergun, D.I. Automated acquisition and analysis of small angle X-ray scattering data. Nuc. Instr. Meth. Phys. Res. Sec. A 689, 52–59 (2012).
Toft, K.N. et al. High-throughput small angle X-ray scattering from proteins in solution using a microfluidic front-end. Anal. Chem. 80, 3648–3654 (2008).
Blanchet, C.E. et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 48, 431–443 (2015).
Petoukhov, M.V. & Svergun, D.I. Applications of small-angle X-ray scattering to biomacromolecular solutions. Int. J. Biochem. Cell Biol. 45, 429–437 (2013).
Koch, M.H., Vachette, P. & Svergun, D.I. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev. Biophys. 36, 147–227 (2003).
Wall, M.E., Gallagher, S.C. & Trewhella, J. Large-scale shape changes in proteins and macromolecular complexes. Ann. Rev. Phys. Chem. 51, 355–380 (2000).
Mertens, H.D. & Svergun, D.I. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 172, 128–141 (2010).
Rambo, R.P. & Tainer, J.A. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr. Opin. Struct. Biol. 20, 128–137 (2010).
Krueger, S., Shin, J.H., Raghunandan, S., Curtis, J.E. & Kelman, Z. Atomistic ensemble modeling and small-angle neutron scattering of intrinsically disordered protein complexes: applied to minichromosome maintenance protein. Biophys. J. 101, 2999–3007 (2011).
Whitten, A.E. et al. The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J. Mol. Biol. 368, 407–420 (2007).
Vestergaard, B. et al. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 5, e134 (2007).
Valentini, E., Kikhney, A.G., Previtali, G., Jeffries, C.M. & Svergun, D.I. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 43, D357–D363 (2015).
Franke, D. & Svergun, D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009).
Svergun, D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999).
Svergun, D.I., Petoukhov, M.V. & Koch, M.H. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 (2001).
Petoukhov, M.V. & Svergun, D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89, 1237–1250 (2005).
Sali, A. et al. Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Structure 23, 1156–1167 (2015).
Panjkovich, A. & Svergun, D.I. Deciphering conformational transitions of proteins by small angle X-ray scattering and normal mode analysis. Phys. Chem. Chem. Phys. 18, 5707–5719 (2015).
Hammel, M. Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS). Eur. Biophys. J. 41, 789–799 (2012).
Bernado, P., Mylonas, E., Petoukhov, M.V., Blackledge, M. & Svergun, D.I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem Soc. 129, 5656–5664 (2007).
Wang, Y., Trewhella, J. & Goldenberg, D.P. Small-angle X-ray scattering of reduced ribonuclease A: effects of solution conditions and comparisons with a computational model of unfolded proteins. J. Mol. Biol. 377, 1576–1592 (2008).
Goldenberg, D.P. & Argyle, B. Minimal effects of macromolecular crowding on an intrinsically disordered protein: a small-angle neutron scattering study. Biophys. J. 106, 905–914 (2014).
Tria, G., Mertens, H.D., Kachala, M. & Svergun, D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2, 207–217 (2015).
Kikhney, A.G. & Svergun, D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 589, 2570–2577 (2015).
Kachala, M., Valentini, E. & Svergun, D.I. Application of SAXS for the Structural Characterization of IDPs. Adv. Exp. Med. Biol. 870, 261–289 (2015).
Jimenez-Garcia, B., Pons, C., Svergun, D.I., Bernado, P. & Fernandez-Recio, J. pyDockSAXS: protein-protein complex structure by SAXS and computational docking. Nucleic Acids Res. 43, W356–W361 (2015).
Cho, H.S., Schotte, F., Dashdorj, N., Kyndt, J. & Anfinrud, P.A. Probing anisotropic structure changes in proteins with picosecond time-resolved small-angle X-ray scattering. J. Phys. Chem. B 117, 15825–15832 (2013).
Graceffa, R. et al. Sub-millisecond time-resolved SAXS using a continuous-flow mixer and X-ray microbeam. J. Synchrotron Radiat. 20, 820–825 (2013).
Roessle, M. et al. Time-resolved small angle scattering: kinetics and structural data from proteins in solution. J. Appl. Crystallogr. 33, 548–551 (2000).
Jacques, D.A. & Trewhella, J. Small-angle scattering for structural biology--expanding the frontier while avoiding the pitfalls. Protein Sci. 19, 642–657 (2010).
Grishaev, A. Sample preparation, data collection and preliminary data analysis in biomolecular solution X-ray scattering. Curr. Protoc. Protein Sci. 70, 17.14.1–17.14.18 (2012).
Glatter, O. & Kratky, O. Small Angle X-ray Scattering (Academic Press, 1982).
Feigin, L.A. & Svergun, D.I. Structure Analysis by Small-angle X-ray and Neutron Scattering (Plenum Press, 1987).
Svergun, D.I., Koch, M.H.J., Timmins, P.A. & May, R.P. Small Angle X-Ray and Neutron Scattering From Solutions of Biological Macromolecules. 1st edn, (Oxford University Press, 2013).
Jacques, D.A., Guss, J.M., Svergun, D.I. & Trewhella, J. Publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution. Acta Crystallogr. Sec. D68, 620–626 (2012).
Svergun, D.I. & Nierhaus, K.H. A map of protein-rRNA distribution in the 70 S Escherichia coli ribosome. J. Biol. Chem. 275, 14432–14439 (2000).
Whitten, A.E., Jeffries, C.M., Harris, S.P. & Trewhella, J. Cardiac myosin-binding protein C decorates F-actin: implications for cardiac function. Proc. Natl. Acad. Sci. USA 105, 18360–18365 (2008).
Lakey, J.H. Neutrons for biologists: a beginner's guide, or why you should consider using neutrons. J. R. Soc. 6, S567–S573 (2009).
Zaccai, G. Straight lines of neutron scattering in biology: a review of basic controls in SANS and EINS. Eur. Biophys. J. 41, 781–787 (2012).
Krueger, S. SANS provides unique information on the structure and function of biological macromolecules in solution. Physica B 241-243, 1131–1137 (1997).
Heller, W.T. & Littrell, K.C. Small-angle neutron scattering for molecular biology: basics and instrumentation. Meth. Mol. Biol. 544, 293–305 (2009).
Krueger, J.K. & Wignall, G.D. Small-Angle Neutron Scattering from Biological Molecules. in Neutron Scattering in Biology 127–160 (Berlin/Heidelberg: Springer, 2006).
Svergun, D.I. & Koch, M.H.J. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 66, 1735–1782 (2003).
Jacrot, B. & Zaccai, G. Determination of molecular weight by neutron scattering. Biopolymers 20, 2413–2426 (1981).
Sears, V.F. Neutron scattering lengths and cross sections. Neutron News 3, 26–37 (1992).
Whitten, A.E., Cai, S. & Trewhella, J. MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J. App. Crystallogr. 41, 222–226 (2008).
Konarev, P.V. & Svergun, D.I. A posteriori determination of the useful data range for small-angle scattering experiments on dilute monodisperse systems. IUCrJ 2, 352–360 (2015).
Lafleur, J.P. et al. Automated microfluidic sample-preparation platform for high-throughput structural investigation of proteins by small-angle X-ray scattering. J. App. Crystallogr. 44, 1090–1099 (2011).
Skou, M., Skou, S., Jensen, T.G., Vestergaard, B. & Gillilan, R.E. icrofluidic dialysis for biological small-angle X-ray scattering. J. Appl. Crystallogr. 47, 1355–1366 (2014).
Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 (1951).
Kuwamoto, S., Akiyama, S. & Fujisawa, T. Radiation damage to a protein solution, detected by synchrotron X-ray small-angle scattering: dose-related considerations and suppression by cryoprotectants. J. Synchrotron Radiat. 11, 462–468 (2004).
Jeffries, C.M., Graewert, M.A., Svergun, D.I. & Blanchet, C.E. Limiting radiation damage for high brilliance biological solution scattering: practical experience at the EMBL P12 beamline, PETRAIII. J. Synchrotron Radiat. 22, 273–279 (2014).
Folta-Stogniew, E. & Williams, K.R. Determination of molecular masses of proteins in solution: implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Tech. 10, 51–63 (1999).
Jacques, D.A. et al. Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state. Acta Crystallogr. D Biol. Crystallogr. 65, 574–581 (2009).
Dünweg, B., Reith, D., Steinhauser, M. & Kremer, K. Corrections to scaling in the hydrodynamic properties of dilute polymer solutions. J. Chem. Phys. 117, 914–924 (2002).
Brewer, A.K. & Striegel, A.M. Characterizing the size, shape, and compactness of a polydisperse prolate ellipsoidal particle via quadruple-detector hydrodynamic chromatography. Analyst 136, 515–519 (2011).
Rubinson, K.A., Stanley, C. & Krueger, S. Small-angle neutron scattering and the errors in protein structures that arise from uncorrected background and intermolecular interactions. J. App. Crystallogr. 41, 456–465 (2008).
Trewhella, J. et al. Report of the wwPDB Small-Angle Scattering Task Force: data requirements for biomolecular modeling and the PDB. Structure 21, 875–881 (2013).
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Gasteiger, E. et al. Protein identification and analysis tools on the ExPASy server. in The Proteomics Protocols Handbook (ed. Walker, J.M.) 571–607 (Humana Press, 2005).
Cleland,, W.W. Dithiothreitol, a new protective reagent for sh groups. Biochemistry 3, 480–482 (1964).
Stevens, R., Stevens, L. & Price, N.C. The stabilities of various thiol compounds used in protein purifications. Biochem. Educ. 11, 70–70 (1983).
Kreżel, A., Latajka, R., Bujacz, G.D. & Bal, W. Coordination properties of Tris(2-carboxyethyl)phosphine, a newly introduced thiol reductant, and its oxide. Inorg. Chem. 42, 1994–2003 (2003).
Krezel, A. et al. Coordination of heavy metals by dithiothreitol, a commonly used thiol group protectant. J. Inorg. Biochem. 84, 77–88 (2001).
Han, J.C. & Han, G.Y. A procedure for quantitative determination of Tris(2-Carboxyethyl)phosphine, an odorless reducing agent more stable and effective than dithiothreitol. Anal. Biochem. 220, 5–10 (1994).
Zhao, H., Brown, P.H. & Schuck, P. On the distribution of protein refractive index increments. Biophys. J. 100, 2309–2317 (2011).
Guinier, A. La diffraction des rayons X aux très pétits angles; application á l'etude de phénomènes ultramicroscopiques. Ann. Phys. 12, 161–237 (1939).
Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992).
Glatter, O. A new method for the evaluation of small-angle scattering data. J. Appl. Crystallogr. 10, 415–421 (1977).
Mylonas, E. & Svergun, D.I. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J. Appl. Crystallogr. 40, s245–s249 (2007).
Krigbaum, W.R. & Kugler, F.R. Molecular conformation of egg-white lysozyme and bovine alpha-lactalbumin in solution. Biochemistry 9, 1216–1223 (1970).
Watson, M.C. & Curtis, J.E. Probing the average local structure of biomolecules using small-angle scattering and scaling laws. Biophys. J. 106, 2474–2482 (2014).
Fischer, H., de Oliveira Neto, M., Napolitano, H.B., Polikarpov, I. & Craievich, A.F. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 43, 101–109 (2010).
Rambo, R.P. & Tainer, J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013).
Fritz, G. & Bergmann, A. SAXS instruments with slit collimation: investigation of resolution and flux. J. Appl. Crystallogr. 39, 64–71 (2006).
Svergun, D.I., Barberato, C. & Koch, M.H.J. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995).
Voss, N.R. & Gerstein, M. Calculation of standard atomic volumes for RNA and comparison with proteins: RNA is packed more tightly. J. Mol. Biol. 346, 477–492 (2005).
Orthaber, D., Bergmann, A. & Glatter, O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Crystallogr. 33, 218–225 (2000).
Wignall, G.D. & Bates, F.S. Absolute calibration of small-angle neutron scattering data. J. App. Crystallogr. 20, 28–40 (1987).
Svergun, D.I. et al. Protein hydration in solution: experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. USA 95, 2267–2272 (1998).
Leiting, B., Marsilio, F. & O'Connell, J.F. Predictable deuteration of recombinant proteins expressed in Escherichia coli. Anal. Biochem. 265, 351–355 (1998).
Goryunov, A.S. H/D isotope effects on protein hydration and interaction in solution. Gen. Physiol. Biophys. 25, 303–311 (2006).
Sheu, S.Y., Schlag, E.W., Selzle, H.L. & Yang, D.Y. Molecular dynamics of hydrogen bonds in protein-D2O: the solvent isotope effect. J. Phys. Chem. A 112, 797–802 (2008).
Baghurst, P.A., Nichol, L.W. & Sawyer, W.H. The effect of D 2 O on the association of -lactoglobulin A. J. Biol. Chem. 247, 3199–3204 (1972).
Dougan, L., Koti, A.S., Genchev, G., Lu, H. & Fernandez, J.M. A single-molecule perspective on the role of solvent hydrogen bonds in protein folding and chemical reactions. Chemphyschem 9, 2836–2847 (2008).
Tehei, M., Madern, D., Pfister, C. & Zaccai, G. Fast dynamics of halophilic malate dehydrogenase and BSA measured by neutron scattering under various solvent conditions influencing protein stability. Proc. Natl. Acad. Sci. USA 98, 14356–14361 (2001).
Jasnin, M., Tehei, M., Moulin, M., Haertlein, M. & Zaccai, G. Solvent isotope effect on macromolecular dynamics in E. coli. Eur. Biophys. J. 37, 613–617 (2008).
Graewert, M.A. et al. Automated pipeline for purification, biophysical and x-ray analysis of biomacromolecular solutions. Sci. Rep. 5, 10734 (2015).
Glasoe, P.K. & Long, F.A. Use of glass electrodes to measure acidities in deuterium oxide. J. Phys. Chem. 64, 188–190 (1960).
Svergun, D.I., Koch, M.H.J., Timmins, P.A. & May, R.P. Small Angle X-Ray and Neutron Scattering from Solutions of Biological Macromolecules. (Oxford University Press, 2013).
Wood, K. et al. Exploring the structure of biological macromolecules in solution using Quokka, the small angle neutron scattering instrument, at ANSTO. Nucl. Instrum. Meth. A 798, 44–51 (2015).
Mokbel, N. et al. K7del is a common TPM2 gene mutation associated with nemaline myopathy and raised myofibre calcium sensitivity. Brain 136, 494–507 (2013).
Mathew, E., Mirza, A. & Menhart, N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J. Synchrotron. Radiat. 11, 314–318 (2004).
David, G. & Perez, J. Combined sampler robot and high-performance liquid chromatography: a fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J. App. Crystallogr. 42, 892–900 (2009).
Chen, X. et al. High yield expression and efficient purification of deuterated human protein galectin-2. Food Bioprod. Process. 90, 563–572 (2012).
Sun, G. et al. Expression, purification and crystallization of human kynurenine aminotransferase 2 exploiting a highly optimized codon set. Protein Expr. Purif. 121, 41–45 (2016).
Acknowledgements
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF) project BIOSCAT, grant 05K12YE1, by the European Community' Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement no. 283570) and by an HFSP grant (RGP0017/2012 to D.I.S. and C.M.J.). M.A.G. was supported by an EMBL Interdisciplinary Postdoc Programme (EIPOD) and Marie Curie COFUND actions. We thank J. Trewhella, in whose laboratory many of the procedures were performed. We also thank A. Duff (Australian Nuclear Science and Technology Organisation (ANSTO)) and David Jacques (LMB, Cambridge) for constructive comments on the deuteration incorporation spreadsheet.
Author information
Authors and Affiliations
Contributions
C.M.J., M.A.G., C.E.B., D.B.L., A.E.W. and D.I.S. helped develop SAXS and SANS sample preparation protocols and analytical tools. C.M.J., M.A.G., C.E.B. and D.I.S. performed radiation damage studies and developed protocols for SEC–SAXS. C.M.J., A.E.W. and D.B.L. contributed to 'in-house' 2H-labeling protocols. D.B.L., A.E.W., C.M.J. and D.I.S. optimized protocols for preparing samples for SANS with contrast variation. A.E.W. developed Contrast. C.M.J., M.A.G., C.E.B., D.B.L., A.E.W. and D.I.S. critically discussed and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests. Although particular commercial products are noted throughout the text, such references should not be interpreted as product endorsements.
Supplementary information
Supplementary Method 1
Calculating molecular weight from absolute scaled data (XLSX 178 kb)
Supplementary Method 2
Calculating 2H labeling of a protein (XLSX 18 kb)
Supplementary Data
Contrast module input file (TXT 0 kb)
Rights and permissions
About this article
Cite this article
Jeffries, C., Graewert, M., Blanchet, C. et al. Preparing monodisperse macromolecular samples for successful biological small-angle X-ray and neutron-scattering experiments. Nat Protoc 11, 2122–2153 (2016). https://doi.org/10.1038/nprot.2016.113
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.113
This article is cited by
-
Quantitative size-resolved characterization of mRNA nanoparticles by in-line coupling of asymmetrical-flow field-flow fractionation with small angle X-ray scattering
Scientific Reports (2023)
-
Human cystatin C induces the disaggregation process of selected amyloid beta peptides: a structural and kinetic view
Scientific Reports (2023)
-
Enabling Efficient Design of Biological Formulations Through Advanced Characterization
Pharmaceutical Research (2023)
-
Realizing the AF4-UV-SAXS on-line coupling on protein and antibodies using high flux synchrotron radiation at the CoSAXS beamline, MAX IV
Analytical and Bioanalytical Chemistry (2023)
-
A comparative study of unpasteurized and pasteurized frozen whole hen eggs using size-exclusion chromatography and small-angle X-ray scattering
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.