Abstract
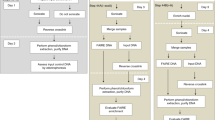
To understand physiological phenomena at the tissue level, elucidation of tissue-specific molecular functions in vivo is required. As an example of the current state of affairs, many genes in plants have been reported to have discordant levels of expression between bulk tissues and the specific tissues in which the respective gene product is principally functional. The principal challenge in deciphering such tissue-specific functions lies in separating tissues with high spatiotemporal resolution to evaluate accurate gene expression profiles. Here, we provide a simple and rapid tissue isolation protocol to isolate all three major leaf tissues (mesophyll, vasculature and epidermis) from Arabidopsis within 30 min with high purity. On the basis of the different cell-to-cell connectivities of tissues, the mesophyll isolation is achieved by making protoplasts, and the vasculature and epidermis isolation is achieved through sonication and enzymatic digestion of leaves. We have successfully tested the protocol on several other plant species, including crop plants such as soybean, tomato and wheat. Furthermore, isolated tissues can be used not only for tissue-specific transcriptome assays but also potentially for tissue-specific proteome and methylome assays.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Endo, M., Shimizu, H., Nohales, A.M., Araki, T. & Kay, S.A. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515, 419–422 (2014).
Shimizu, H. et al. Decentralized clock discretely process thermal and photoperiodic cues in a specific tissue. Nat. Plants 1, 15163 (2015).
Endo, M., Nakamura, S., Araki, T., Mochizuki, N. & Nagatani, A. Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17, 1941–1952 (2005).
Endo, M., Mochizuki, N., Suzuki, T. & Nagatani, A. CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19, 84–93 (2007).
Kozuka, T., Kong, S.G., Doi, M., Shimazaki, K. & Nagatani, A. Tissue-autonomous promotion of palisade cell development by phototropin 2 in Arabidopsis. Plant Cell 23, 3684–3695 (2011).
Ranjan, A., Fiene, G., Fackendahl, P. & Hoecker, U. The Arabidopsis repressor of light signaling SPA1 acts in the phloem to regulate seedling de-etiolation, leaf expansion and flowering time. Development 138, 1851–1862 (2011).
Kleber, R. & Kehr, J. Preparation and quality assessment of RNA from cell-specific samples obtained by laser microdissection. Methods Mol. Biol. 323, 367–377 (2006).
Birnbaum, K. et al. A gene expression map of the Arabidopsis root. Science 302, 1956–1960 (2003).
Brady, S.M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007).
Birnbaum, K. et al. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2, 615–619 (2005).
Michael, T.P. et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4, e14 (2008).
Fukuda, H., Nakamichi, N., Hisatsune, M., Murase, H. & Mizuno, T. Synchronization of plant circadian oscillators with a phase delay effect of the vein network. Phys. Rev. Lett. 99, 098102 (2007).
Wenden, B., Toner, D.L., Hodge, S.K., Grima, R. & Millar, A.J. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc. Natl. Acad. Sci. USA 109, 6757–6762 (2012).
Muranaka, T., Kubota, S. & Oyama, T. A single-cell bioluminescence imaging system for monitoring cellular gene expression in a plant body. Plant Cell Physiol. 54, 2085–2093 (2013).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002).
Miura, F., Enomoto, Y., Dairiki, R. & Ito, T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012).
Sun, L. et al. Ultrasensitive and fast bottom-up analysis of femtogram amounts of complex proteome digests. Angew. Chem. Int. Ed. Engl. 52, 13661–13664 (2013).
Huang, E.L. et al. SNaPP: simplified nano-proteomics platform for reproducible global proteomic analysis of nanogram protein quantities. Endocrinology 157, 1307–1314 (2016).
Acknowledgements
We thank T. Koto for technical assistance and J.A. Hejna for English proofreading. This work was supported by JST PRESTO grant 888067 (to M.E.), JSPS KAKENHI grants 16H01240, 25650097 and 15H05958 (to M.E.), grants from the Nakatani Foundation (to M.E.) and the Mitsubishi Foundation (to M.E.), and Grants-in-Aid for Scientific Research on Priority Areas 19060012 and 19060016 (to T.A.).
Author information
Authors and Affiliations
Contributions
M.E. and H.S. conceived and designed the protocol and experimental procedures. M.E. wrote the manuscript. M.E. and T.A. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Isolated mesophyll, vasculature, and epidermis from crop plants.
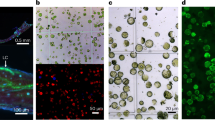
Isolated mesophyll (a, d, g), vasculature (b, e, h), and epidermis (c, f, i) from a cotyledon or primary leaf of soybean (a-c), tomato (d-f) and wheat (g-i). Bars = 250 µm.
Supplementary Figure 2 RNA purification and cDNA amplification from other crops.
Semi-quantitative RT-PCR detection for beta-tubulin genes in leaf tissue of Arabidopsis, tomato, soybean, and wheat. Non-RT controls were run to confirm that there was no false positive amplification from genomic DNA. W; whole leaf, M; mesophyll, V; vasculature, E; epidermis.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 (PDF 379 kb)
Rights and permissions
About this article
Cite this article
Endo, M., Shimizu, H. & Araki, T. Rapid and simple isolation of vascular, epidermal and mesophyll cells from plant leaf tissue. Nat Protoc 11, 1388–1395 (2016). https://doi.org/10.1038/nprot.2016.083
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.083
This article is cited by
-
Conservation of dynamic characteristics of transcriptional regulatory elements in periodic biological processes
BMC Bioinformatics (2022)
-
Leaf layer-based transcriptome profiling for discovery of epidermal-selective promoters in Medicago truncatula
Planta (2022)
-
Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in Arabidopsis thaliana
Nature Communications (2021)
-
The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway
Nature Communications (2020)
-
Post-translational modifications drive plant cell differentiation
Plant Cell, Tissue and Organ Culture (PCTOC) (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.