Abstract
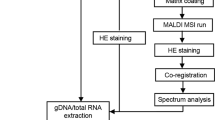
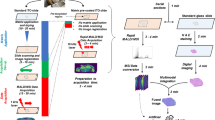
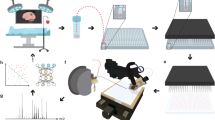
Formalin-fixed and paraffin-embedded (FFPE) tissue specimens are the gold standard for histological examination, and they provide valuable molecular information in tissue-based research. Metabolite assessment from archived tissue samples has not been extensively conducted because of a lack of appropriate protocols and concerns about changes in metabolite content or chemical state due to tissue processing. We present a protocol for the in situ analysis of metabolite content from FFPE samples using a high-mass-resolution matrix-assisted laser desorption/ionization fourier-transform ion cyclotron resonance mass spectrometry imaging (MALDI-FT-ICR-MSI) platform. The method involves FFPE tissue sections that undergo deparaffinization and matrix coating by 9-aminoacridine before MALDI-MSI. Using this platform, we previously detected ∼1,500 m/z species in the mass range m/z 50–1,000 in FFPE samples; the overlap compared with fresh frozen samples is 72% of m/z species, indicating that metabolites are largely conserved in FFPE tissue samples. This protocol can be reproducibly performed on FFPE tissues, including small samples such as tissue microarrays and biopsies. The procedure can be completed in a day, depending on the size of the sample measured and raster size used. Advantages of this approach include easy sample handling, reproducibility, high throughput and the ability to demonstrate molecular spatial distributions in situ. The data acquired with this protocol can be used in research and clinical practice.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wishart, D.S. Proteomics and the human metabolome project. Expert Rev. Proteomics 4, 333–335 (2007).
Fiehn, O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002).
Cacciatore, S. & Loda, M. Innovation in metabolomics to improve personalized healthcare. Ann. N. Y. Acad. Sci. 1346, 57–62 (2015).
Nilsson, A. et al. Mass spectrometry imaging in drug development. Anal. Chem. 87, 1437–1455 (2015).
Buck, A. et al. High-resolution MALDI-FT-ICR MS imaging for the analysis of metabolites from formalin-fixed paraffin-embedded clinical tissue samples. J. Pathol. 237, 123–132 (2015).
Fan, T.W., Lane, A.N., Higashi, R.M. & Yan, J. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics 7, 257–269 (2011).
Engskog, M. et al. Metabolic profiling of epithelial ovarian cancer cell lines: evaluation of harvesting protocols for profiling using NMR spectroscopy. Bioanalysis 7, 157–166 (2015).
Cesare Marincola, F., Dessi, A., Corbu, S., Reali, A. & Fanos, V. Clinical impact of human breast milk metabolomics. Clin. Chim. Acta 451, 103–106 (2015).
Chadeau-Hyam, M. et al. Meeting-in-the-middle using metabolic profiling - a strategy for the identification of intermediate biomarkers in cohort studies. Biomarkers 16, 83–88 (2011).
Emwas, A.H. et al. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: a review. Metabolomics 11, 872–894 (2015).
Yang, Y. et al. Study of metabonomic profiles of human esophageal carcinoma by use of high-resolution magic-angle spinning 1H NMR spectroscopy and multivariate data analysis. Anal. Bioanal. Chem. 405, 3381–3389 (2013).
Lutz, N.W., Beraud, E. & Cozzone, P.J. Metabolomic analysis of rat brain by high resolution nuclear magnetic resonance spectroscopy of tissue extracts. J. Vis. Exp. 91, 51829 (2014).
Macura, S., Mishra, P.K., Gamez, J.D. & Pirko, I. MR microscopy of formalin fixed paraffin embedded histology specimens. Magn. Reson. Med. 71, 1989–1994 (2014).
Liu, J. et al. Outcome-related metabolomic patterns from 1H/31P NMR after mild hypothermia treatments of oxygen-glucose deprivation in a neonatal brain slice model of asphyxia. J. Cereb. Blood Flow Metab. 31, 547–559 (2011).
Noren, B. et al. Absolute quantification of human liver metabolite concentrations by localized in vivo 31P NMR spectroscopy in diffuse liver disease. Eur. Radiol. 15, 148–157 (2005).
Baker, M.J. et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 9, 1771–1791 (2014).
Ellis, D.I. & Goodacre, R. Metabolic fingerprinting in disease diagnosis: biomedical applications of infrared and Raman spectroscopy. Analyst 131, 875–885 (2006).
Lasch, P., Chiriboga, L., Yee, H. & Diem, M. Infrared spectroscopy of human cells and tissue: detection of disease. Technol. Cancer Res. Treat. 1, 1–7 (2002).
Liu, Q. & Xiao, S. Effects of spectral resolution and signal-to-noise ratio of hyperspectral sensors on retrieving atmospheric parameters. Opt. Lett. 39, 60–63 (2014).
Wang, L. & Mizaikoff, B. Application of multivariate data-analysis techniques to biomedical diagnostics based on mid-infrared spectroscopy. Anal. Bioanal. Chem. 391, 1641–1654 (2008).
Junot, C., Fenaille, F., Colsch, B. & Becher, F. High resolution mass spectrometry based techniques at the crossroads of metabolic pathways. Mass Spectrom. Rev. 33, 471–500 (2014).
Hayasaka, T. et al. Matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight (MALDI-QIT-TOF)-based imaging mass spectrometry reveals a layered distribution of phospholipid molecular species in the mouse retina. Rapid Commun. Mass Spectrom. 22, 3415–3426 (2008).
Sun, N. et al. High-resolution metabolite imaging of light and dark treated retina using MALDI-FTICR mass spectrometry. Proteomics 14, 913–923 (2014).
Liu, N.Q. et al. Proteomics pipeline for biomarker discovery of laser capture microdissected breast cancer tissue. J. Mammary Gland Biol. Neoplasia 17, 155–164 (2012).
Umar, A., Luider, T.M., Foekens, J.A. & Pasa-Tolic, L. NanoLC-FT-ICR MS improves proteome coverage attainable for approximately 3000 laser-microdissected breast carcinoma cells. Proteomics 7, 323–329 (2007).
Norris, J.L. & Caprioli, R.M. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 113, 2309–2342 (2013).
Balluff, B. et al. MALDI imaging identifies prognostic seven-protein signature of novel tissue markers in intestinal-type gastric cancer. Am. J. Pathol. 179, 2720–2729 (2011).
Cohen, S.L. & Chait, B.T. Influence of matrix solution conditions on the MALDI-MS analysis of peptides and proteins. Anal. Chem. 68, 31–37 (1996).
Powers, T.W. et al. MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PloS One 9, e106255 (2014).
Mitchell, C.A., Long, H., Donaldson, M., Francese, S. & Clench, M.R. Lipid changes within the epidermis of living skin equivalents observed across a time-course by MALDI-MS imaging and profiling. Lipids Health Dis. 14, 84 (2015).
Huber, K. et al. A rapid ex vivo tissue model for optimising drug detection and ionisation in MALDI imaging studies. Histochem. Cell Biol. 142, 361–371 (2014).
Aichler, M. et al. Spatially resolved quantification of gadolinium(III)-based magnetic resonance agents in tissue by MALDI imaging mass spectrometry after in vivo MRI. Angew. Chem. Int. Ed. Engl. 54, 4279–4283 (2015).
Elsner, M. et al. MALDI imaging mass spectrometry reveals COX7A2, TAGLN2 and S100-A10 as novel prognostic markers in Barrett's adenocarcinoma. J. Proteomics 75, 4693–4704 (2012).
Lazova, R., Seeley, E.H., Keenan, M., Gueorguieva, R. & Caprioli, R.M. Imaging mass spectrometry—a new and promising method to differentiate Spitz nevi from Spitzoid malignant melanomas. Am. J. Dermatopathol. 34, 82–90 (2012).
Anderson, D.M. et al. High resolution MALDI imaging mass spectrometry of retinal tissue lipids. J. Am. Soc. Mass Spectrom. 25, 1394–1403 (2014).
Grey, A.C. & Schey, K.L. Age-related changes in the spatial distribution of human lens alpha-crystallin products by MALDI imaging mass spectrometry. Invest. Ophthalmol. Vis. Sci. 50, 4319–4329 (2009).
Zemski Berry, K.A., Gordon, W.C., Murphy, R.C. & Bazan, N.G. Spatial organization of lipids in the human retina and optic nerve by MALDI imaging mass spectrometry. J. Lipid Res. 55, 504–515 (2014).
Calligaris, D. et al. MALDI mass spectrometry imaging analysis of pituitary adenomas for near-real-time tumor delineation. Proc. Natl. Acad. Sci. USA 112, 9978–9983 (2015).
Rompp, A. & Spengler, B. Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 139, 759–783 (2013).
Miura, D., Fujimura, Y. & Wariishi, H. In situ metabolomic mass spectrometry imaging: recent advances and difficulties. J. Proteomics 75, 5052–5060 (2012).
Dekker, T.J. et al. Towards imaging metabolic pathways in tissues. Anal. Bioanal. Chem. 407, 2167–2176 (2015).
Rompp, A. et al. Histology by mass spectrometry: label-free tissue characterization obtained from high-accuracy bioanalytical imaging. Angew. Chem. Int. Ed. Engl. 49, 3834–3838 (2010).
Blum, F. Der formaldehyd als haertungsmittel. Z Wiss Mikrosk 10, 314–315 (1893).
Amalou, H. & Wood, B.J. Biopsy and personalized medicine. Nat. Rev. Gastroenterol. Hepatol. 9, 683 (2012).
Travis, W.D., Brambilla, E., Burke, A.P., Marx, A. & Nicholson, A.G. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J. Thorac. Oncol. 10, 1240–1242 (2015).
Sobin, L.H., Gospodarowicz, M.K. & Wittekind, C. TNM Classification of Malignant Tumours (Wiley, 2011).
Berg, D. et al. Protein microarray-based comparison of HER2, estrogen receptor, and progesterone receptor status in core biopsies and surgical specimens from FFPE breast cancer tissues. Appl. Immunohistochem. Mol. Morphol. 19, 300–305 (2011).
Kononen, J. et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 4, 844–847 (1998).
Leong, T.Y., Cooper, K. & Leong, A.S. Immunohistology—past, present, and future. Adv. Anat. Pathol. 17, 404–418 (2010).
Auerbach, S.S. et al. RNA-Seq-based toxicogenomic assessment of fresh frozen and formalin-fixed tissues yields similar mechanistic insights. J. Appl. Toxicol. 35, 766–780 (2014).
Heydt, C. et al. Comparison of pre-analytical FFPE sample preparation methods and their impact on massively parallel sequencing in routine diagnostics. PloS One 9, e104566 (2014).
Specht, K., Richter, T., Muller, U., Walch, A. & Hofler, M.W. Quantitative gene expression analysis in microdissected archival tissue by real-time RT-PCR. J. Mol. Med. (Berl) 78, B27 (2000).
Fanelli, M., Amatori, S., Barozzi, I. & Minucci, S. Chromatin immunoprecipitation and high-throughput sequencing from paraffin-embedded pathology tissue. Nat. Protoc. 6, 1905–1919 (2011).
Hedegaard, J. et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PloS One 9, e98187 (2014).
Azimzadeh, O. et al. Formalin-fixed paraffin-embedded (FFPE) proteome analysis using gel-free and gel-based proteomics. J. Proteome Res. 9, 4710–4720 (2010).
Magdeldin, S. & Yamamoto, T. Toward deciphering proteomes of formalin-fixed paraffin-embedded (FFPE) tissues. Proteomics 12, 1045–1058 (2012).
Wisniewski, J.R. Proteomic sample preparation from formalin fixed and paraffin embedded tissue. J. Vis. Exp. http://dx.doi.org/10.3791/50589 (2013).
Casadonte, R. & Caprioli, R.M. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat. Protoc. 6, 1695–1709 (2011).
Casadonte, R. et al. Imaging mass spectrometry to discriminate breast from pancreatic cancer metastasis in formalin-fixed paraffin-embedded tissues. Proteomics 14, 956–964 (2014).
Gustafsson, O.J. et al. MALDI imaging mass spectrometry of N-linked glycans on formalin-fixed paraffin-embedded murine kidney. Anal. Bioanal. Chem. 407, 2127–2139 (2015).
Casadonte, R. et al. Imaging mass spectrometry analysis of renal amyloidosis biopsies reveals protein co-localization with amyloid deposits. Anal. Bioanal. Chem. 407, 5323–5331 (2015).
Wei, Q., Xiao, X., Fogle, P. & Dong, Z. Changes in metabolic profiles during acute kidney injury and recovery following ischemia/reperfusion. PloS One 9, e106647 (2014).
Jove, M., Portero-Otin, M., Naudi, A., Ferrer, I. & Pamplona, R. Metabolomics of human brain aging and age-related neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 73, 640–657 (2014).
Warburg, O. On the origin of cancer cells. Science 123, 309–314 (1956).
Renshaw, S. Immunochemical Staining Techniques chapter 4 (Scion Publishing, 2007).
Kelly, A.D. et al. Metabolomic profiling from formalin-fixed, paraffin-embedded tumor tissue using targeted LC/MS/MS: application in sarcoma. PloS One 6, e25357 (2011).
Yuan, M., Breitkopf, S.B., Yang, X. & Asara, J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 7, 872–881 (2012).
Wojakowska, A. et al. An optimized method of metabolite extraction from formalin-fixed paraffin-embedded tissue for GC/MS analysis. PloS One 10, e0136902 (2015).
Wojakowska, A. et al. Detection of metabolites discriminating subtypes of thyroid cancer: molecular profiling of FFPE samples using the GC/MS approach. Mol. Cell. Endocrinol. 417, 149–157 (2015).
Sahm, F. et al. Detection of 2-hydroxyglutarate in formalin-fixed paraffin-embedded glioma specimens by gas chromatography/mass spectrometry. Brain Pathol. 22, 26–31 (2012).
Bruinen, A.L. et al. Mass spectrometry imaging of drug related crystal-like structures in formalin-fixed frozen and paraffin-embedded rabbit kidney tissue sections. J. Am. Soc. Mass Spectrom. 27, 117–123 (2016).
Ng, E.S., Kangarloo, S.B., Konno, M., Paterson, A. & Magliocco, A.M. Extraction of tamoxifen and its metabolites from formalin-fixed, paraffin-embedded tissues: an innovative quantitation method using liquid chromatography and tandem mass spectrometry. Cancer Chemother. Pharmacol. 73, 475–484 (2014).
Moreno-Gordaliza, E. et al. Elemental bioimaging in kidney by LA-ICP-MS as a tool to study nephrotoxicity and renal protective strategies in cisplatin therapies. Anal. Chem. 83, 7933–7940 (2011).
Wang, J. et al. MALDI-TOF MS imaging of metabolites with a N-(1-naphthyl) ethylenediamine dihydrochloride matrix and its application to colorectal cancer liver metastasis. Anal. Chem. 87, 422–430 (2015).
Thomas, A., Charbonneau, J.L., Fournaise, E. & Chaurand, P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal. Chem. 84, 2048–2054 (2012).
Korte, A.R. & Lee, Y.J. MALDI-MS analysis and imaging of small molecule metabolites with 1,5-diaminonaphthalene (DAN). J. Mass Spectrom. 49, 737–741 (2014).
Diehl, H.C. et al. The challenge of on-tissue digestion for MALDI MSI- a comparison of different protocols to improve imaging experiments. Anal. Bioanal. Chem. 407, 2223–2243 (2015).
Hughes, C., Gaunt, L., Brown, M., Clarke, N.W. & Gardner, P. Assessment of paraffin removal from prostate FFPE sections using transmission mode FTIR-FPA imaging. Anal. Methods 6, 1028–1035 (2014).
Buck, A. et al. How suitable is MALDI-TOF for metabolite imaging from clinical formalin-fixed and paraffin-embedded tissue samples in comparison to MALDI-FT-ICR mass spectrometry? Anal. Chem. 88, 5281–5289 (2016).
Goodwin, R.J. et al. Qualitative and quantitative MALDI imaging of the positron emission tomography ligands raclopride (a D2 dopamine antagonist) and SCH 23390 (a D1 dopamine antagonist) in rat brain tissue sections using a solvent-free dry matrix application method. Anal. Chem. 83, 9694–9701 (2011).
Groseclose, M.R. & Castellino, S. A mimetic tissue model for the quantification of drug distributions by MALDI imaging mass spectrometry. Anal. Chem. 85, 10099–10106 (2013).
Pirman, D.A. & Yost, R.A. Quantitative tandem mass spectrometric imaging of endogenous acetyl-L-carnitine from piglet brain tissue using an internal standard. Anal. Chem. 83, 8575–8581 (2011).
McDonnell, L.A. et al. Peptide and protein imaging mass spectrometry in cancer research. J. Proteomics 73, 1921–1944 (2010).
Qi, L.W., Wang, C.Z. & Yuan, C.S. Isolation and analysis of ginseng: advances and challenges. Nat. Prod. Rep. 28, 467–495 (2011).
Ceglarek, U. et al. Challenges and developments in tandem mass spectrometry based clinical metabolomics. Mol. Cell. Endocrinol. 301, 266–271 (2009).
Vinaixa, M. et al. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: state of the field and future prospects. Trends Anal. Chem 78, 23–35.
Kaletas, B.K. Sample preparation issues for tissue imaging by imaging MS. Proteomics 9, 2622–2633 (2009).
Yang, J. & Caprioli, R.M. Matrix pre-coated targets for high throughput MALDI imaging of proteins. J. Mass Spectrom. 49, 417–422 (2014).
Vermillion-Salsbury, R.L. & Hercules, D.M. 9-Aminoacridine as a matrix for negative mode matrix-assisted laser desorption/ionization. Rapid Commun. Mass Spectrom. 16, 1575–1581 (2002).
Sun, G. et al. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal. Chem. 80, 7576–7585 (2008).
Miura, D. et al. Ultrahighly sensitive in situ metabolomic imaging for visualizing spatiotemporal metabolic behaviors. Anal. Chem. 82, 9789–9796 (2010).
Yukihira, D. et al. MALDI efficiency of metabolites quantitatively associated with their structural properties: a quantitative structure-property relationship (QSPR) approach. J. Am. Soc. Mass Spectrom. 25, 1–5 (2014).
Yang, J. & Caprioli, R.M. Matrix precoated targets for direct lipid analysis and imaging of tissue. Anal. Chem. 85, 2907–2912 (2013).
Goodwin, R.J. Sample preparation for mass spectrometry imaging: small mistakes can lead to big consequences. J. Proteomics 75, 4893–4911 (2012).
Thomas, A. & Chaurand, P. Advances in tissue section preparation for MALDI imaging MS. Bioanalysis 6, 967–982 (2014).
Smith, C.A. et al. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751 (2005).
Wishart, D.S. et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 41, D801–D807 (2013).
Fahy, E., Sud, M., Cotter, D. & Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35, W606–W612 (2007).
Pence, H.E. & Williams, A. ChemSpider: an online chemical information resource. J. Chem. Educ. 87, 1123–1124 (2010).
Gemperline, E. & Li, L. MALDI-mass spectrometric imaging for the investigation of metabolites in Medicago truncatula root nodules. J. Vis. Exp. http://dx.doi.org/10.3791/51434 (2014).
Xia, J. & Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 6, 743–760 (2011).
Gundisch, S. et al. Critical roles of specimen type and temperature before and during fixation in the detection of phosphoproteins in breast cancer tissues. Lab. Invest. 95, 561–571 (2015).
Deininger, S.O. et al. Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Anal. Bioanal. Chem. 401, 167–181 (2011).
Fonville, J.M. et al. Robust data processing and normalization strategy for MALDI mass spectrometric imaging. Anal. Chem. 84, 1310–1319 (2012).
Alexandrov, T. MALDI imaging mass spectrometry: statistical data analysis and current computational challenges. BMC Bioinformatics 13, 1–13 (2012).
McDonnell, L.A., van Remoortere, A., de Velde, N., van Zeijl, R.J. & Deelder, A.M. Imaging mass spectrometry data reduction: automated feature identification and extraction. J. Am. Soc. Mass Spectrom. 21, 1969–1978 (2010).
Mantini, D. et al. LIMPIC: a computational method for the separation of protein MALDI-TOF-MS signals from noise. BMC Bioinformatics 8, 101 (2007).
Bodger, K., Campbell, F. & Rhodes, J.M. Detection of sulfated glycoproteins in intestinal metaplasia: a comparison of traditional mucin staining with immunohistochemistry for the sulfo-Lewis(a) carbohydrate epitope. J. Clin. Pathol. 56, 703–708 (2003).
Endo, T. et al. Expression of sulfated carbohydrate chain and core peptides of mucin detected by monoclonal antibodies in Barrett's esophagus and esophageal adenocarcinoma. J. Gastroenterol. 33, 811–815 (1998).
Spengler, B. Mass spectrometry imaging of biomolecular information. Anal. Chem. 87, 64–82 (2015).
Acknowledgements
This project was funded by the Ministry of Education and Research of the Federal Republic of Germany (BMBF; grant number 01ZX1310B), ERA-NET TRANSCAN-2-ARREST and the Deutsche Forschungsgemeinschaft (grant nos. HO 1254/3-1, SFB 824 TP Z02 and WA 1656/3-1). The authors thank U. Buchholz, C.-M. Pflüger, G. Mettenleiter and A. Voss for technical assistance, and D. Borgmann for bioinformatics assistance.
Author information
Authors and Affiliations
Contributions
A.L. and A.B. were responsible for the MALDI imaging mass spectrometry data acquisition and data analyses, and wrote the manuscript. B.B. was involved in data and bioinformatics development, and wrote the manuscript. N.S. and K.G. conducted protocol validation experiments. A.F. and K.-P.J. assisted in interpretation of the results. K.-P.J., P.J.K.K., C.J.H.v.d.V., G.W., F.E., R.L. and M. Aubele provided samples and assisted in histological interpretation of the samples. L.M. was involved in data and bioinformatics development. H.Z. and M. Aichler assisted in interpretation of the results and writing of the manuscript. A.W. conceived the study, and assisted in interpretation of results and writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ly, A., Buck, A., Balluff, B. et al. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat Protoc 11, 1428–1443 (2016). https://doi.org/10.1038/nprot.2016.081
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.081
This article is cited by
-
Metabolic heterogeneity affects trastuzumab response and survival in HER2-positive advanced gastric cancer
British Journal of Cancer (2024)
-
Mass spectrometry-based proteomics as an emerging tool in clinical laboratories
Clinical Proteomics (2023)
-
A pilot radiometabolomics integration study for the characterization of renal oncocytic neoplasia
Scientific Reports (2023)
-
Low-melting point agarose as embedding medium for MALDI mass spectrometry imaging and laser-capture microdissection-based proteomics
Scientific Reports (2023)
-
Spatial metabolomics identifies distinct tumor-specific and stroma-specific subtypes in patients with lung squamous cell carcinoma
npj Precision Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.