Abstract
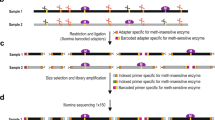
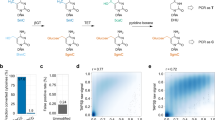
Active DNA demethylation is mediated by ten-eleven translocation (TET) proteins that progressively oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). We have developed a methylation-assisted bisulfite sequencing (MAB-seq) method that enables direct genome-scale mapping and quantification of 5fC and 5caC marks together at single-base resolution. In bisulfite sequencing (BS), unmethylated cytosine residues (Cs), 5fCs and 5caCs, are converted to uracil and cannot be discriminated from each other. The pretreatment of the DNA with the CpG methylation enzyme M.SssI, which converts only the Cs to 5mCs, protects Cs but not 5fCs and 5caCs, which enables direct detection of 5fCs and 5caCs as uracils. Here we also describe an adapted version of the protocol to perform reduced-representation MAB-seq (RRMAB-seq) that provides increased coverage on CpG-rich regions, thus reducing the execution costs and increasing the feasibility of the technique. The main advantage of MAB-seq is to reduce the number of chemical/enzymatic DNA treatments required before bisulfite treatment and to avoid the need for prohibitive sequencing coverage, thus making it more reliable and affordable than subtractive approaches. The method presented here is the ideal tool for studying DNA demethylation dynamics in any biological system. Overall timing is ∼3 d for library preparation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402 (2000).
Reik, W. & Walter, J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2, 21–32 (2001).
Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662–673 (2002).
Okano, M., Bell, D.W., Haber, D.A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Smith, Z.D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Neri, F. et al. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell 155, 121–134 (2013).
Rhee, I. et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416, 552–556 (2002).
Kohli, R.M. & Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
Neri, F. et al. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 14, R91 (2013).
Neri, F. et al. TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene 34, 4168–4176 (2015).
Neri, F. et al. TET1 is controlled by pluripotency-associated factors in ESCs and downmodulated by PRC2 in differentiated cells and tissues. Nucleic Acids Res. 43, 6814–6826 (2015).
Cortázar, D. et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423 (2011).
He, Y.F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Spruijt, C.G. et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152, 1146–1159 (2013).
Iurlaro, M. et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 14, R119 (2013).
Bachman, M. et al. 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 11, 555–557 (2015).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
Harris, R.A. et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 28, 1097–1105 (2010).
Gu, H. et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution 7, 133–136 (2010).
Huang, Y. et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 5, e8888 (2010).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Neri, F. et al. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep. 10, 674–683 (2015).
Xu, M., Kladde, M.P., Van Etten, J.L. & Simpson, R.T. Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res. 26, 3961–3966 (1998).
Booth, M.J., Marsico, G., Bachman, M., Beraldi, D. & Balasubramanian, S. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nat. Chem. 6, 435–440 (2014).
Jin, S.G., KAdam, S. & Pfeifer, G.P. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 38, e125 (2010).
Holmes, E.E. et al. Performance evaluation of kits for bisulfite-conversion of DNA from tissues, cell lines, FFPE tissues, aspirates, lavages, effusions, plasma, serum, and urine. PLoS ONE 9, e93933 (2014).
Song, C.-X. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 (2013).
Booth, M.J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012).
Raiber, E.-A. et al. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymineDNA glycosylase. Genome Biol. 13, R69 (2012).
Shen, L. et al. Genome-wide analysis revealsTET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706 (2013).
Lu, X. et al. Chemical modification-assisted bisulfite sequencing (CAB-Seq) for 5-carboxylcytosine detection in DNA. J. Am. Chem. Soc. 135, 9315–9317 (2013).
Incarnato, D., Krepelova, A. & Neri, F. High-throughput single nucleotide variant discovery in E14 mouse embryonic stem cells provides a new reference genome assembly. Genomics 104, 121–127 (2014).
Xi, Y. & Li, W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009).
Acknowledgements
This work was supported by the Associazione Italiana Ricerca sul Cancro (AIRC) IG 2011 11982 to S.O.
Author information
Authors and Affiliations
Contributions
F.N. conceived the MAB-seq. D.I. and F.N. developed the protocol and the computational methods, and analyzed the data. A.K. and C.P. contributed to the MAB optimization. S.O. supervised the experiments. F.N., D.I. and S.O. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data 1 — Bsmap_postprocess (step 99).
This custom perl script elaborates the output SAM file derived from Bsmap software and it generates two independent files: a SAM file of the mapped reads and a FastQ file of the unmapped reads. (TXT 3 kb)
Supplementary Data 2 — 5fcaC_bintest (step 104).
This custom perl script elaborates the methratio file generated in step 103 and estimates the significance (p-value) of each cytosine to be 5fC/5caC taking into account the M.SssI methylation fail rate calculated in step 102. (TXT 3 kb)
Rights and permissions
About this article
Cite this article
Neri, F., Incarnato, D., Krepelova, A. et al. Methylation-assisted bisulfite sequencing to simultaneously map 5fC and 5caC on a genome-wide scale for DNA demethylation analysis. Nat Protoc 11, 1191–1205 (2016). https://doi.org/10.1038/nprot.2016.063
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.063
This article is cited by
-
Distinctive epigenomes characterize glioma stem cells and their response to differentiation cues
Genome Biology (2018)
-
DNA methylation profiles in cancer diagnosis and therapeutics
Clinical and Experimental Medicine (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.