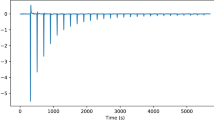
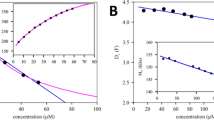
Abstract
Isothermal titration calorimetry (ITC) is a powerful and widely used method to measure the energetics of macromolecular interactions by recording a thermogram of differential heating power during a titration. However, traditional ITC analysis is limited by stochastic thermogram noise and by the limited information content of a single titration experiment. Here we present a protocol for bias-free thermogram integration based on automated shape analysis of the injection peaks, followed by combination of isotherms from different calorimetric titration experiments into a global analysis, statistical analysis of binding parameters and graphical presentation of the results. This is performed using the integrated public-domain software packages NITPIC, SEDPHAT and GUSSI. The recently developed low-noise thermogram integration approach and global analysis allow for more precise parameter estimates and more reliable quantification of multisite and multicomponent cooperative and competitive interactions. Titration experiments typically take 1–2.5 h each, and global analysis usually takes 10–20 min.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Robinson, C.V., Sali, A. & Baumeister, W. The molecular sociology of the cell. Nature 450, 973–982 (2007).
Cebecauer, M., Spitaler, M., Sergé, A. & Magee, A.I. Signalling complexes and clusters: functional advantages and methodological hurdles. J. Cell Sci. 123, 309–320 (2010).
Ladbury, J.E. & Arold, S.T. Noise in cellular signaling pathways: causes and effects. Trends Biochem. Sci. 37, 173–178 (2012).
Ladbury, J.E., Klebe, G. & Freire, E. Adding calorimetric data to decision making in lead discovery: a hot tip. Nat. Rev. Drug Discov. 9, 23–27 (2010).
Chaires, J.B. Calorimetry and thermodynamics in drug design. Annu. Rev. Biophys. 37, 135–151 (2008).
Freire, E., Mayorga, O.L. & Straume, M. Isothermal titration calorimetry. Anal. Chem. 62, 950A–959A (1990).
Wiseman, T., Williston, S., Brandts, J.F. & Lin, L.N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 (1989).
Cooper, A. Microcalorimetry of protein-protein interactions. Methods Mol. Biol. 88, 11–22 (1998).
Willerich, I. & Gröhn, F. Molecular structure encodes nanoscale assemblies: understanding driving forces in electrostatic self-assembly. J. Am. Chem. Soc. 133, 20341–20356 (2011).
Heerklotz, H., Tsamaloukas, A.D. & Keller, S. Monitoring detergent-mediated solubilization and reconstitution of lipid membranes by isothermal titration calorimetry. Nat. Protoc. 4, 686–697 (2009).
Tsamaloukas, A.D., Keller, S. & Heerklotz, H. Uptake and release protocol for assessing membrane binding and permeation by way of isothermal titration calorimetry. Nat. Protoc. 2, 695–704 (2007).
Bains, G. & Freire, E. Calorimetric determination of cooperative interactions in high affinity binding processes. Anal. Biochem. 192, 203–206 (1991).
Brown, A. Analysis of cooperativity by isothermal titration calorimetry. Int. J. Mol. Sci. 10, 3457–3477 (2009).
Freire, E., Schön, A. & Velázquez-Campoy, A. Isothermal titration calorimetry: general formalism using binding polynomials. Methods Enzymol. 455, 127–155 (2009).
Houtman, J.C.D. et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 (2007).
Baker, B.M. & Murphy, K.P. Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophys. J. 71, 2049–2055 (1996).
Coussens, N.P., Schuck, P. & Zhao, H. Strategies for assessing proton linkage to bimolecular interactions by global analysis of isothermal titration calorimetry data. J. Chem. Thermodyn. 52, 95–107 (2012).
Keller, S. et al. High-precision isothermal titration calorimetry with automated peak shape analysis. Anal. Chem. 84, 5066–5073 (2012).
Pierce, M.M., Raman, C.S. & Nall, B.T. Isothermal titration calorimetry of protein-protein interactions. Methods 19, 213–221 (1999).
Velázquez-Campoy, A. & Freire, E. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat. Protoc. 1, 186–191 (2006).
Roselin, L.S., Lin, M.-S., Lin, P.-H., Chang, Y. & Chen, W.-Y. Recent trends and some applications of isothermal titration calorimetry in biotechnology. Biotechnol. J. 5, 85–98 (2010).
Velázquez-Campoy, A., Ohtaka, H., Nezami, A., Muzammil, S. & Freire, E. Isothermal titration calorimetry. Curr. Protoc. Cell Biol. 17.8, 17.8.1–17.8.24 (2004).
Torres, F.E., Recht, M.I., Coyle, J.E., Bruce, R.H. & Williams, G. Higher throughput calorimetry: opportunities, approaches and challenges. Curr. Opin. Struct. Biol. 20, 598–605 (2010).
Scheuermann, T.H. & Brautigam, C.A. High-precision, automated integration of multiple isothermal titration calorimetric thermograms: new features of NITPIC. Methods 76, 87–98 (2015).
Herman, P. & Lee, J.C. Functional energetic landscape in the allosteric regulation of muscle pyruvate kinase. 1. Calorimetric study. Biochemistry 48, 9448–9455 (2009).
Krishnamoorthy, J. & Mohanty, S. Open-ITC: an alternate computational approach to analyze the isothermal titration calorimetry data of complex binding mechanisms. J. Mol. Recognit. 24, 1056–1066 (2011).
Henzl, M.T. Characterization of parvalbumin and polcalcin divalent ion binding by isothermal titration calorimetry. Methods Enzymol. 455, 259–297 (2009).
Freiburger, L.A., Auclair, K. & Mittermaier, A.K. Elucidating protein binding mechanisms by variable-c ITC. Chembiochem 10, 2871–2873 (2009).
Schönbeck, C., Holm, R. & Westh, P. Higher order inclusion complexes and secondary interactions studied by global analysis of calorimetric titrations. Anal. Chem. 84, 2305–2312 (2012).
Zhao, H., Piszczek, G. & Schuck, P. SEDPHAT—A platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods 76, 137–148 (2015).
Armstrong, K.M. & Baker, B.M. A comprehensive calorimetric investigation of an entropically driven T cell receptor-peptide/major histocompatibility complex interaction. Biophys. J. 93, 597–609 (2007).
Freiburger, L.A. et al. Competing allosteric mechanisms modulate substrate binding in a dimeric enzyme. Nat. Struct. Mol. Biol. 18, 288–294 (2011).
Herman, P. & Lee, J.C. Functional energetic landscape in the allosteric regulation of muscle pyruvate kinase. 2. Fluorescence study. Biochemistry 48, 9456–9465 (2009).
Zhao, H. & Schuck, P. Global multi-method analysis of affinities and cooperativity in complex systems of macromolecular interactions. Anal. Chem. 84, 9513–9519 (2012).
Zhao, H. & Schuck, P. Combining biophysical methods for the analysis of protein complex stoichiometry and affinity in SEDPHAT. Acta Crystallogr. D Biol. Crystallogr. 71, 3–14 (2015).
Freiburger, L., Auclair, K. & Mittermaier, A. Global ITC fitting methods in studies of protein allostery. Methods 76, 149–161 (2015).
Houtman, J.C.D. et al. Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat. Struct. Mol. Biol. 13, 798–805 (2006).
Wynn, R.M., Li, J., Brautigam, C.A., Chuang, J.L. & Chuang, D.T. Structural and biochemical characterization of human mitochondrial branched-chain α-ketoacid dehydrogenase phosphatase. J. Biol. Chem. 287, 9178–9192 (2012).
Duff, M.R., Grubbs, J., Serpersu, E. & Howell, E.E. Weak interactions between folate and osmolytes in solution. Biochemistry 51, 2309–2318 (2012).
Huang, L., Serganov, A. & Patel, D.J. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol. Cell 40, 774–786 (2010).
Gustchina, E. et al. Complexes of neutralizing and non-neutralizing affinity matured Fabs with a mimetic of the internal trimeric coiled-coil of HIV-1 gp41. PLoS One 8, e78187 (2013).
Dellarole, M., Sánchez, I.E. & de Prat Gay, G. Thermodynamics of cooperative DNA recognition at a replication origin and transcription regulatory site. Biochemistry 49, 10277–10286 (2010).
Krainer, G., Broecker, J., Vargas, C., Fanghänel, J. & Keller, S. Quantifying high-affinity binding of hydrophobic ligands by isothermal titration calorimetry. Anal. Chem. 84, 10715–10722 (2012).
Moncrieffe, M.C., Grossmann, J.G. & Gay, N.J. Assembly of oligomeric death domain complexes during Toll receptor signaling. J. Biol. Chem. 283, 33447–33454 (2008).
Muscroft-Taylor, A.C., Soares da Costa, T.P. & Gerrard, J.A. New insights into the mechanism of dihydrodipicolinate synthase using isothermal titration calorimetry. Biochimie 92, 254–262 (2010).
Pellizzaro, M.L. et al. Conformer-independent ureidoimidazole motifs—tools to probe conformational and tautomeric effects on the molecular recognition of triply hydrogen-bonded heterodimers. Chem. Eur. J. 17, 14508–14517 (2011).
Krainer, G. & Keller, S. Single-experiment displacement assay for quantifying high-affinity binding by isothermal titration calorimetry. Methods 76, 116–123 (2015).
Brautigam, C.A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015).
Baranauskienė, L. & Matulis, D. Intrinsic thermodynamics of ethoxzolamide inhibitor binding to human carbonic anhydrase XIII. BMC Biophys. 5, 12 (2012).
Matulis, D. & Todd, M. in Biocalorimetry 2: Applications of Calorimetry in the Biological Sciences (eds. Ladbury, J.E. & Doyle, M.E.), 107–132 (John Wiley & Sons, 2004).
Broecker, J., Vargas, C. & Keller, S. Revisiting the optimal c value for isothermal titration calorimetry. Anal. Biochem. 418, 307–309 (2011).
Gans, P., Sabatini, A. & Vacca, A. Simultaneous calculation of equilibrium constants and standard formation enthalpies from calorimetric data for systems with multiple equilibria in solution. J. Solution Chem. 37, 467–476 (2008).
Brautigam, C.A. Fitting two- and three-site binding models to isothermal titration calorimetric data. Methods 76, 124–136 (2015).
Vega, S., Abian, O. & Velázquez-Campoy, A. A unified framework based on the binding polynomial for characterizing biological systems by isothermal titration calorimetry. Methods 76, 99–115 (2015).
Herrera, I. & Winnik, M. Differential binding models for isothermal titration calorimetry: moving beyond the Wiseman isotherm. J. Phys. Chem. B 117, 8659–8672 (2013).
Minetti, C., Privalov, P.L. & Remeta, D.P. in Proteins in Solution and at Interfaces: Methods and Applications in Biotechnology and Materials Science (eds. Ruso, J.M. & Pineiro, A.), 139–179 (Wiley, 2013).
Goldberg, R., Kishore, N. & Lennen, R.M. Thermodynamic quantities for the ionization reactions of buffers. J. Phys. Chem. Ref. Data 31, 231–370 (1999).
Bevington, P.R. & Robinson, D.K. Data Reduction and Error Analysis for the Physical Sciences (Mc-Graw-Hill, 1992).
Textor, M., Vargas, C. & Keller, S. Calorimetric quantification of linked equilibria in cyclodextrin/lipid/detergent mixtures for membrane-protein reconstitution. Methods 76, 183–193 (2015).
Textor, M. & Keller, S. Calorimetric quantification of cyclodextrin-mediated detergent extraction for membrane-protein reconstitution. Methods Enzymol. 567, 129–156 (2016).
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through International Research Training Group 1830, the Stiftung Rheinland–Pfalz für Innovation and the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering, US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
C.V. and S.K. collected data. C.A.B., H.Z., C.V., S.K. and P.S. performed data analysis. C.A.B., H.Z., C.V., S.K. and P.S. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Screenshot of the SEDPHAT window after importing the data from NITPIC.
Shown are the isotherms from the different binding experiments, each in a separate graphics with subpanels for the raw data (top, circles and error bars) and the residuals (bottom). Initially, the residuals are meaningless due to the lack of a fitting model. In each experiment graph, a set of pushbuttons in the upper right corner provides access to individual experimental parameters (blue numbered button) and functions for data management (small buttons labeled i and x to inactivate or delete the titration). To get a quick overview, the identity of each titration along with local experimental parameters can be printed as text over the respective panels by pressing control-O.
Supplementary Figure 2 Screenshot of the SEDPHAT window after the fit has converged.
In each experiment panel, the best-fit titration model is now drawn as a solid line. It should be noted that the residuals are now on a reasonable scale, and for most experiments lack significant systematicity. The overlay of textual output reporting global and local fit parameters can be toggled on and off with the keys ESC and control-O. The best-fit parameters can be obtained also in the model parameter box (control-P), or be summarized in a displayed ASCII text-file (control-T).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 (PDF 460 kb)
Supplementary Data
Example Data Sets from the duplicate titration experiments of CAII with TFMSA in five different buffers. (ZIP 168 kb)
Rights and permissions
About this article
Cite this article
Brautigam, C., Zhao, H., Vargas, C. et al. Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions. Nat Protoc 11, 882–894 (2016). https://doi.org/10.1038/nprot.2016.044
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.044
This article is cited by
-
Isothermal titration calorimetry
Nature Reviews Methods Primers (2023)
-
Structural maturation of SYCP1-mediated meiotic chromosome synapsis by SYCE3
Nature Structural & Molecular Biology (2023)
-
Engineering of bidirectional, cyanobacteriochrome-based light-inducible dimers (BICYCL)s
Nature Methods (2023)
-
Synergistic activation of the insulin receptor via two distinct sites
Nature Structural & Molecular Biology (2022)
-
Multivalent binding kinetics resolved by fluorescence proximity sensing
Communications Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.