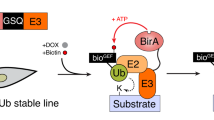
Abstract
Ubiquitination is an essential protein modification that influences eukaryotic processes ranging from substrate degradation to nonproteolytic pathway alterations, including DNA repair and endocytosis. Previous attempts to analyze substrates via physical association with their respective ubiquitin ligases have had some success. However, because of the transient nature of enzyme-substrate interactions and rapid protein degradation, detection of substrates remains a challenge. Ligase trapping is an affinity purification approach in which ubiquitin ligases are fused to a polyubiquitin-binding domain, which allows the isolation of ubiquitinated substrates. Immunoprecipitation is first used to enrich for proteins that are bound to the ligase trap. Subsequently, affinity purification is used under denaturing conditions to capture proteins conjugated with hexahistidine-tagged ubiquitin. By using this protocol, ubiquitinated substrates that are specific for a given ligase can be isolated for mass spectrometry or western blot analysis. After cells have been collected, the described protocol can be completed in 2–3 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Breitschopf, K., Bengal, E., Ziv, T., Admon, A. & Ciechanover, A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17, 5964–5973 (1998).
Schulman, B.A. Twists and turns in ubiquitin-like protein conjugation cascades. Protein Sci. 20, 1941–1954 (2011).
Finley, D., Ulrich, H.D., Sommer, T. & Kaiser, P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360 (2012).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Benanti, J.A., Cheung, S.K., Brady, M.C. & Toczyski, D.P. A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat. Cell Biol. 9, 1184–1191 (2007).
Yen, H.C. & Elledge, S.J. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322, 923–929 (2008).
Yen, H.C., Xu, Q., Chou, D.M., Zhao, Z. & Elledge, S.J. Global protein stability profiling in mammalian cells. Science 322, 918–923 (2008).
Emanuele, M.J. et al. Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 (2011).
Barral, Y., Jentsch, S. & Mann, C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 9, 399–409 (1995).
Koepp, D.M., Kile, A.C., Swaminathan, S. & Rodriguez-Rivera, V. The F-box protein Dia2 regulates DNA replication. Mol. Biol. Cell 17, 1540–1548 (2006).
Landry, B.D., Doyle, J.P., Toczyski, D.P. & Benanti, J.A. F-box protein specificity for G1 cyclins is dictated by subcellular localization. PLoS Genet. 8, e1002851 (2012).
Pant, V. & Lozano, G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev. 28, 1739–1751 (2014).
Galligan, J.T. et al. Proteomic analysis and identification of cellular interactors of the giant ubiquitin ligase HERC2. J. Proteome Res. 14, 953–966 (2015).
Martinez-Noel, G. et al. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol. Cell Biol. 32, 3095–3106 (2012).
Davis, M.A. et al. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with mediator. Genes Dev. 27, 151–156 (2013).
Pierce, N.W., Kleiger, G., Shan, S.O. & Deshaies, R.J. Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 (2009).
Gardner, R.G., Shearer, A.G. & Hampton, R.Y. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol. Cell Biol. 21, 4276–4291 (2001).
Tagwerker, C. et al. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell Proteomics 5, 737–748 (2006).
Elsasser, S. & Finley, D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 7, 742–749 (2005).
Grabbe, C. & Dikic, I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 109, 1481–1494 (2009).
Sims, J.J., Haririnia, A., Dickinson, B.C., Fushman, D. & Cohen, R.E. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat. Struct. Mol. Biol. 16, 883–889 (2009).
Mark, K.G., Simonetta, M., Maiolica, A., Seller, C.A. & Toczyski, D.P. Ubiquitin ligase trapping identifies an SCFSaf1 pathway targeting unprocessed vacuolar/lysosomal proteins. Mol. Cell 53, 148–161 (2014).
Bennett, E.J., Rush, J., Gygi, S.P. & Harper, J.W. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143, 951–965 (2010).
Kuchay, S. et al. FBXL2- and PTPL1-mediated degradation of p110-free p85β regulatory subunit controls the PI(3)K signalling cascade. Nat. Cell Biol. 15, 472–480 (2013).
Wimuttisuk, W. et al. Novel Cul3 binding proteins function to remodel E3 ligase complexes. BMC Cell Biol. 15, 28 (2014).
Peng, J. et al. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 (2003).
Gururaja, T., Li, W., Noble, W.S., Payan, D.G. & Anderson, D.C. Multiple functional categories of proteins identified in an in vitro cellular ubiquitin affinity extract using shotgun peptide sequencing. J. Proteome Res. 2, 394–404 (2003).
Hitchcock, A.L., Auld, K., Gygi, S.P. & Silver, P.A. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA 100, 12735–12740 (2003).
Kirkpatrick, D.S., Weldon, S.F., Tsaprailis, G., Liebler, D.C. & Gandolfi, A.J. Proteomic identification of ubiquitinated proteins from human cells expressing His-tagged ubiquitin. Proteomics 5, 2104–2111 (2005).
Jeon, H.B. et al. A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochem. Biophys. Res. Commun. 357, 731–736 (2007).
Meierhofer, D., Wang, X., Huang, L. & Kaiser, P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J. Proteome Res. 7, 4566–4576 (2008).
Franco, M., Seyfried, N.T., Brand, A.H., Peng, J. & Mayor, U. A novel strategy to isolate ubiquitin conjugates reveals wide role for ubiquitination during neural development. Mol. Cell Proteomics http://dx.doi.org/10.1074/mcp.M110.002188 (2011).
Danielsen, J.M. et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell Proteomics 10 http://dx.doi.org/10.1074/mcp.M110.003590 (2011).
Wilson, M.D., Saponaro, M., Leidl, M.A. & Svejstrup, J.Q. MultiDsk: a ubiquitin-specific affinity resin. PLoS One 7, e46398 (2012).
Layfield, R. et al. Purification of poly-ubiquitinated proteins by S5a-affinity chromatography. Proteomics 1, 773–777 (2001).
Anindya, R., Aygun, O. & Svejstrup, J.Q. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell 28, 386–397 (2007).
Hjerpe, R. et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 (2009).
Shi, Y. et al. A data set of human endogenous protein ubiquitination sites. Mol. Cell Proteomics 10 http://dx.doi.org/10.1074/mcp.M110.002089 (2011).
Lopitz-Otsoa, F. et al. Integrative analysis of the ubiquitin proteome isolated using Tandem Ubiquitin Binding Entities (TUBEs). J. Proteomics 75, 2998–3014 (2012).
Tan, M.K., Lim, H.J., Bennett, E.J., Shi, Y. & Harper, J.W. Parallel SCF adaptor capture proteomics reveals a role for SCFFBXL17 in NRF2 activation via BACH1 repressor turnover. Mol. Cell 52, 9–24 (2013).
Behrends, C., Sowa, M.E., Gygi, S.P. & Harper, J.W. Network organization of the human autophagy system. Nature 466, 68–76 (2010).
Sowa, M.E., Bennett, E.J., Gygi, S.P. & Harper, J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009).
Loveless, T.B. et al. DNA damage regulates translation through β-TRCP targeting of CReP. PLoS Genet. 11, e1005292 (2015).
Kim, T.Y. et al. Substrate trapping proteomics reveals targets of the β-TrCP2/FBXW11 ubiquitin ligase. Mol. Cell Biol. 35, 167–181 (2015).
Peng, J. & Gygi, S.P. Proteomics: the move to mixtures. J. Mass Spectrom. 36, 1083–1091 (2001).
Xu, G., Paige, J.S. & Jaffrey, S.R. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 (2010).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Theurillat, J.P. et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 346, 85–89 (2014).
Kronke, J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014).
Thompson, J.W. et al. Quantitative Lys-ɛ-Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 ligase HUWE1. J. Biol. Chem. 289, 28942–28955 (2014).
Ordureau, A., Munch, C. & Harper, J.W. Quantifying ubiquitin signaling. Mol. Cell 58, 660–676 (2015).
Sultan, M. et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321, 956–960 (2008).
Kulathu, Y., Akutsu, M., Bremm, A., Hofmann, K. & Komander, D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 16, 1328–1330 (2009).
Rahighi, S. et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109 (2009).
van Wijk, S.J., Muller, S. & Dikic, I. Shared and unique properties of ubiquitin and SUMO interaction networks in DNA repair. Genes Dev. 25, 1763–1769 (2011).
McEwan, D.G. & Dikic, I. The Three Musketeers of Autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 21, 195–201 (2011).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grants R01 GM059691 and GM070539 to D.P.T., and by a predoctoral fellowship from the American Heart Association to T.B.L.
Author information
Authors and Affiliations
Contributions
K.G.M. developed the protocol for yeast application, designed and performed experiments, analyzed data and wrote the manuscript; T.B.L. developed the protocol for mammalian cell application, designed and performed experiments, analyzed data and wrote the manuscript; D.P.T. conceived and oversaw the study, designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Mark, K., Loveless, T. & Toczyski, D. Isolation of ubiquitinated substrates by tandem affinity purification of E3 ligase–polyubiquitin-binding domain fusions (ligase traps). Nat Protoc 11, 291–301 (2016). https://doi.org/10.1038/nprot.2016.008
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.008
This article is cited by
-
Proteomic strategies for characterizing ubiquitin-like modifications
Nature Reviews Methods Primers (2021)
-
Gene expression and cell identity controlled by anaphase-promoting complex
Nature (2020)
-
A substrate-trapping strategy to find E3 ubiquitin ligase substrates identifies Parkin and TRIM28 targets
Communications Biology (2020)
-
Using proteomics to identify ubiquitin ligase–substrate pairs: how novel methods may unveil therapeutic targets for neurodegenerative diseases
Cellular and Molecular Life Sciences (2019)
-
An E2-ubiquitin thioester-driven approach to identify substrates modified with ubiquitin and ubiquitin-like molecules
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.