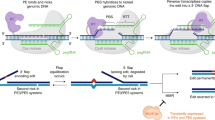
Abstract
Programmable nucleases enable engineering of the genome by utilizing endogenous DNA double-strand break (DSB) repair pathways. Although homologous recombination (HR)-mediated gene knock-in is well established, it cannot necessarily be applied in every cell type and organism because of variable HR frequencies. We recently reported an alternative method of gene knock-in, named the PITCh (Precise Integration into Target Chromosome) system, assisted by microhomology-mediated end-joining (MMEJ). MMEJ harnesses independent machinery from HR, and it requires an extremely short homologous sequence (5–25 bp) for DSB repair, resulting in precise gene knock-in with a more easily constructed donor vector. Here we describe a streamlined protocol for PITCh knock-in, including the design and construction of the PITCh vectors, and their delivery to either human cell lines by transfection or to frog embryos by microinjection. The construction of the PITCh vectors requires only a few days, and the entire process takes ∼1.5 months to establish knocked-in cells or ∼1 week from injection to early genotyping in frog embryos.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Sakuma, T. & Woltjen, K. Nuclease-mediated genome editing: at the front-line of functional genomics technology. Dev. Growth Differ. 56, 2–13 (2014).
Bibikova, M., Golic, M., Golic, K.G. & Carroll, D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161, 1169–1175 (2002).
Lee, H.J., Kim, E. & Kim, J.S. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 20, 81–89 (2010).
Lee, H.J., Kweon, J., Kim, E., Kim, S. & Kim, J.S. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 22, 539–548 (2012).
Taleei, R. & Nikjoo, H. Biochemical DSB-repair model for mammalian cells in G1 and early S phases of the cell cycle. Mutat. Res. 756, 206–212 (2013).
Hockemeyer, D. et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27, 851–857 (2009).
Hockemeyer, D. et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734 (2011).
Sommer, D. et al. Efficient genome engineering by targeted homologous recombination in mouse embryos using transcription activator-like effector nucleases. Nat. Commun. 5, 3045 (2014).
Mashimo, T. et al. Efficient gene targeting by TAL effector nucleases coinjected with exonucleases in zygotes. Sci. Rep. 3, 1253 (2013).
Gratz, S.J. et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Yasue, A. et al. Highly efficient targeted mutagenesis in one-cell mouse embryos mediated by the TALEN and CRISPR/Cas systems. Sci. Rep. 4, 5705 (2014).
Peng, D., Kurup, S.P., Yao, P.Y., Minning, T.A. & Tarleton, R.L. CRISPR-Cas9–mediated single-gene and gene family disruption in Trypanosoma cruzi. MBio 6, e02097–14 (2014).
Bae, S., Kweon, J., Kim, H.S. & Kim, J.S. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11, 705–706 (2014).
Li, H.L. et al. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 4, 143–154 (2015).
Nakade, S. et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 5, 5560 (2014).
Cristea, S. et al. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol. Bioeng. 110, 871–880 (2013).
Maresca, M., Lin, V.G., Guo, N. & Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 23, 539–546 (2013).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Sakuma, T. et al. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 18, 315–326 (2013).
Hisano, Y. et al. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 5, 8841 (2015).
Xiong, X. et al. 53BP1 promotes microhomology-mediated end-joining in G1-phase cells. Nucleic Acids Res. 43, 1659–1670 (2015).
McVey, M. RPA puts the brakes on MMEJ. Nat. Struct. Mol. Biol. 21, 348–349 (2014).
Kim, H. & Kim, J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334 (2014).
Li, J., Zhang, B., Bu, J. & Du, J. Intron-based genomic editing: a highly efficient method for generating knock-in zebrafish. Oncotarget 6, 17891–17894 (2015).
Chen, F. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755 (2011).
Bedell, V.M. et al. In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118 (2012).
Meyer, M., Ortiz, O., Hrabé de Angelis, M., Wurst, W. & Kühn, R. Modeling disease mutations by gene targeting in one-cell mouse embryos. Proc. Natl. Acad. Sci. USA 109, 9354–9359 (2012).
Orlando, S.J. et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 38, e152 (2010).
Auer, T.O., Duroure, K., De Cian, A., Concordet, J.P. & Del Bene, F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24, 142–153 (2014).
Sakuma, T. et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci. Rep. 3, 3379 (2013).
Sakuma, T. & Yamamoto, T. Engineering customized TALENs using the Platinum Gate TALEN Kit. Methods Mol. Biol. 1338, 61–70 (2016).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Sakuma, T., Nishikawa, A., Kume, S., Chayama, K. & Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 4, 5400 (2014).
Ochiai, H. et al. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells 15, 875–885 (2010).
Mashiko, D. et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3, 3355 (2013).
Certo, M.T. et al. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods 8, 671–676 (2011).
Guschin, D.Y. et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 649, 247–256 (2010).
Vouillot, L. et al. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 5, 407–415 (2015).
Suzuki, K.T. et al. High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol. Open 2, 448–452 (2013).
Nakagawa, Y. et al. Screening methods to identify TALEN-mediated knockout mice. Exp. Anim. 63, 79–84 (2014).
Ota, S. et al. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells 18, 450–458 (2013).
Dahlem, T.J. et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861 (2012).
Pyzocha, N.K., Ran, F.A., Hsu, P.D. & Zhang, F. RNA-guided genome editing of mammalian cells. Methods Mol. Biol. 1114, 269–277 (2014).
Byrne, S.M., Mali, P. & Church, G.M. Genome editing in human stem cells. Methods Enzymol. 546, 119–138 (2014).
Shaner, N.C. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013).
Zhu, Z., Verma, N., González, F., Shi, Z.D. & Huangfu, D. A CRISPR/Cas-mediated selection-free knock-in strategy in human embryonic stem cells. Stem Cell Reports 4, 1103–1111 (2015).
Rodgers, K. & McVey, M. Error-prone repair of DNA double-strand breaks. J. Cell Physiol. 231, 15–24 (2016).
Sakuma, T. et al. Homologous recombination-independent large gene cassette knock-in in CHO cells using TALEN and MMEJ-directed donor plasmids. Int. J. Mol. Sci. 16, 23849–23866 (2015).
Sive, H., Grainger, R. & Harland, R. Early Development of Xenopus laevis: a Laboratory Manual (Cold Spring Harbor Laboratory Press, 2000).
Acknowledgements
The authors express their appreciation to A. Kawahara and Y. Hisano (University of Yamanashi, Yamanashi, Japan) for co-developing the modified PITCh system. We also thank H. Ochiai (Hiroshima University, Hiroshima, Japan) for sharing the synthesized mNeonGreen cDNA under the license agreement with Allele Biotechnology and Pharmaceuticals, Inc. This work was supported by the Japan Society for the Promotion of Science (25890014 to T.S., 25124708 to K.-I.T.S. and 26290070 to T.Y.), the Sasakawa Foundation (to S.N.), the Uehara Memorial Foundation (to T.S.) and the Ministry of Health, Labor, and Welfare of Japan (to T.Y.).
Author information
Authors and Affiliations
Contributions
T.S. organized and wrote the manuscript. S.N. performed the human cell experiments and wrote the manuscript concerning human cell procedures. Y.S. performed the frog experiments. K.-I.T.S. wrote the manuscript concerning frog procedures. T.Y. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
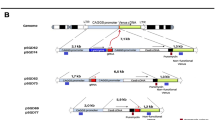
Supplementary Figure 1 Design and validation of the PITCh-gRNAs.
Three gRNAs were initially designed, and safety and efficacy validated by online off-target searches and single-strand annealing (SSA) assays1, respectively. (a) Target sequence of each artificially-designed PITCh-gRNA. PAM sequence is underlined. (b) In silico validation of each PITCh-gRNA using the CRISPR design tool (http://crispr.mit.edu/). Higher scores indicate lower off-target risks in corresponding organisms. (c) Experimental validation of DSB-inducing activities by human cell-based SSA assay. pSTL-ZFA36 vector1 was used for the positive control zinc-finger nuclease (ZFN). Blue bars indicate negative control samples that reporter vectors harboring unrelated sequence were introduced. Red bars indicate test samples that reporter vectors harboring corresponding target sequence were introduced.
1. Ochiai, H. et al. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells 15, 875–885 (2010).
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 (PDF 281 kb)
Rights and permissions
About this article
Cite this article
Sakuma, T., Nakade, S., Sakane, Y. et al. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc 11, 118–133 (2016). https://doi.org/10.1038/nprot.2015.140
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.140
This article is cited by
-
REMOVER-PITCh: microhomology-assisted long-range gene replacement with highly multiplexed CRISPR-Cas9
In Vitro Cellular & Developmental Biology - Animal (2024)
-
Collateral lethality between HDAC1 and HDAC2 exploits cancer-specific NuRD complex vulnerabilities
Nature Structural & Molecular Biology (2023)
-
Viral vectors and extracellular vesicles: innate delivery systems utilized in CRISPR/Cas-mediated cancer therapy
Cancer Gene Therapy (2023)
-
Targeting DNA repair pathways with B02 and Nocodazole small molecules to improve CRIS-PITCh mediated cassette integration in CHO-K1 cells
Scientific Reports (2023)
-
A transient increase of HIF-1α during the G1 phase (G1-HIF) ensures cell survival under nutritional stress
Cell Death & Disease (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.