Abstract
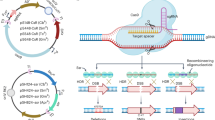
Allelic exchange is an efficient method of bacterial genome engineering. This protocol describes the use of this technique to make gene knockouts and knock-ins, as well as single-nucleotide insertions, deletions and substitutions, in Pseudomonas aeruginosa. Unlike other approaches to allelic exchange, this protocol does not require heterologous recombinases to insert or excise selective markers from the target chromosome. Rather, positive and negative selections are enabled solely by suicide vector–encoded functions and host cell proteins. Here, mutant alleles, which are flanked by regions of homology to the recipient chromosome, are synthesized in vitro and then cloned into allelic exchange vectors using standard procedures. These suicide vectors are then introduced into recipient cells by conjugation. Homologous recombination then results in antibiotic-resistant single-crossover mutants in which the plasmid has integrated site-specifically into the chromosome. Subsequently, unmarked double-crossover mutants are isolated directly using sucrose-mediated counter-selection. This two-step process yields seamless mutations that are precise to a single base pair of DNA. The entire procedure requires ∼2 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Giske, C.G., Monnet, D.L., Cars, O., Carmeli, Y. & ReAct-Action on Antibiotic Resistance Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 52, 813–821 (2008).
Pendleton, J.N., Gorman, S.P. & Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert review of anti-infective therapy 11, 297–308 (2013).
Boucher, H.W. et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 (2009).
Held, K., Ramage, E., Jacobs, M., Gallagher, L. & Manoil, C. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 194, 6387–6389 (2012).
Jacobs, M.A. et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100, 14339–14344 (2003).
Liberati, N.T. et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103, 2833–2838 (2006).
Labaer, J. et al. The Pseudomonas aeruginosa PA01 gene collection. Genome Res. 14, 2190–2200 (2004).
Whiteley, M. et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864 (2001).
Chugani, S. et al. Strain-dependent diversity in the Pseudomonas aeruginosa quorum-sensing regulon. Proc. Natl. Acad. Sci. USA 109, E2823–E2831 (2012).
Wurtzel, O. et al. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 8, e1002945 (2012).
Dotsch, A. et al. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS ONE 7, e31092 (2012).
Gomez-Lozano, M., Marvig, R.L., Molin, S. & Long, K.S. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ. Microbiol. 14, 2006–2016 (2012).
Jones, C.J. et al. ChIP-seq and RNA-seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10, e1003984 (2014).
Blanka, A. et al. Identification of the alternative sigma factor SigX regulon and its implications for Pseudomonas aeruginosa pathogenicity. J. Bacteriol. 196, 345–356 (2014).
Balasubramanian, D. et al. Deep sequencing analyses expands the Pseudomonas aeruginosa AmpR regulon to include small RNA-mediated regulation of iron acquisition, heat shock and oxidative stress response. Nucleic Acids Res. 42, 979–998 (2014).
Gallagher, L.A., Shendure, J. & Manoil, C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2, e00315 (2011).
Winsor, G.L. et al. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600 (2011).
Link, A.J., Phillips, D. & Church, G.M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237 (1997).
Quandt, J. & Hynes, M.F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127, 15–21 (1993).
Hoang, T.T., Karkhoff-Schweizer, R.R., Kutchma, A.J. & Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998).
Ruvkun, G.B. & Ausubel, F.M. A general method for site-directed mutagenesis in prokaryotes. Nature 289, 85–88 (1981).
Goldberg, J.B. & Ohman, D.E. Construction and characterization of Pseudomonas aeruginosa algB mutants: role of algB in high-level production of alginate. J. Bacteriol. 169, 1593–1602 (1987).
Flynn, J.L. & Ohman, D.E. Use of a gene replacement cosmid vector for cloning alginate conversion genes from mucoid and nonmucoid Pseudomonas aeruginosa strains: algS controls expression of algT. J. Bacteriol. 170, 3228–3236 (1988).
Schweizer, H.P. Alielic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6, 1195–1204 (1992).
Quenee, L., Lamotte, D. & Polack, B. Combined sacB-based negative selection and Cre-lox antibiotic marker recycling for efficient gene deletion in Pseudomonas aeruginosa. Biotechniques 38, 63–67 (2005).
Shanks, R.M., Caiazza, N.C., Hinsa, S.M., Toutain, C.M. & O'Toole, G.A. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl. Environ. Microbiol. 72, 5027–5036 (2006).
Choi, K.H. & Schweizer, H.P. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5, 30 (2005).
Tseng, B.S. et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 15, 2865–2878 (2013).
Wolfgang, M.C., Lee, V.T., Gilmore, M.E. & Lory, S. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4, 253–263 (2003).
Fulcher, N.B., Holliday, P.M., Klem, E., Cann, M.J. & Wolfgang, M.C. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol. Microbiol. 76, 889–904 (2010).
Almblad, H. et al. The cyclic AMP-Vfr signaling pathway in Pseudomonas aeruginosa is inhibited by cyclic di-GMP. J. Bacteriol. 197, 2190–2200 (2015).
Zhao, K. et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 497, 388–391 (2013).
Cohen, D. et al. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 112, 11359–11364 (2015).
Colvin, K.M. et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928 (2012).
Colvin, K.M. et al. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7, e1001264 (2011).
Zimmermann, A., Reimmann, C., Galimand, M. & Haas, D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5, 1483–1490 (1991).
Choi, K.H., Kumar, A. & Schweizer, H.P. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells. J. Microbiol. Methods 64, 391–397 (2006).
Campbell, A.M. Episomes. Adv. Genet. 11, 101–145 (1963).
Bochner, B.R., Huang, H.C., Schieven, G.L. & Ames, B.N. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143, 926–933 (1980).
Ried, J.L. & Collmer, A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57, 239–246 (1987).
Schafer, A. et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 (1994).
Pelicic, V., Reyrat, J.M. & Gicquel, B. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on Mycobacteria. J. Bacteriol. 178, 1197–1199 (1996).
Steinmetz, M., Le Coq, D., Djemia, H.B. & Gay, P. Analyse génétique de sacB, gène de structure d'une enzyme secrétée, la lévane-saccharase de Bacillus subtilis Marburg. Mol. Gen. Genet. 191, 138–144 (1983).
Gay, P., Le Coq, D., Steinmetz, M., Berkelman, T. & Kado, C.I. Positive selection procedure for entrapment of insertion sequence elements in Gram-negative bacteria. J. Bacteriol. 164, 918–921 (1985).
Blomfield, I.C., Vaughn, V., Rest, R.F. & Eisenstein, B.I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5, 1447–1457 (1991).
Gregg, C.J. et al. Rational optimization of tolC as a powerful dual selectable marker for genome engineering. Nucleic Acids Res. 42, 4779–4790 (2014).
Jager, W., Schafer, A., Kalinowski, J. & Puhler, A. Isolation of insertion elements from Gram-positive Brevibacterium, Corynebacterium and Rhodococcus strains using the Bacillus subtilis sacB gene as a positive selection marker. FEMS Microbiol. Lett. 126, 1–6 (1995).
Fu, R. & Voordouw, G. Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143 (Part 6): 1815–1826 (1997).
Simon, R., Hotte, B., Klauke, B. & Kosier, B. Isolation and characterization of insertion sequence elements from Gram-negative bacteria by using new broad-host-range, positive selection vectors. J. Bacteriol. 173, 1502–1508 (1991).
Kung, V.L., Ozer, E.A. & Hauser, A.R. The accessory genome of Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 74, 621–641 (2010).
Fowler, R.C. & Hanson, N.D. Emergence of carbapenem resistance due to the novel insertion sequence ISPa8 in Pseudomonas aeruginosa. PLoS ONE 9, e91299 (2014).
Klose, K.E. & Mekalanos, J.J. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28, 501–520 (1998).
Metcalf, W.W. et al. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35, 1–13 (1996).
Barekzi, N. et al. High-frequency Flp recombinase-mediated inversions of the oriC-containing region of the Pseudomonas aeruginosa genome. J. Bacteriol. 182, 7070–7074 (2000).
Choi, K.-H. & Schweizer, H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161 (2006).
Datsenko, K.A. & Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Tischer, B.K., von Einem, J., Kaufer, B. & Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40, 191–197 (2006).
Karlinsey, J.E. lambda-Red genetic engineering in Salmonella enterica serovar typhimurium. Methods Enzymol. 421, 199–209 (2007).
Sharan, S.K., Thomason, L.C., Kuznetsov, S.G. & Court, D.L. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009).
Murphy, K.C., Campellone, K.G. & Poteete, A.R. PCR-mediated gene replacement in Escherichia coli. Gene 246, 321–330 (2000).
Liang, R. & Liu, J. Scarless and sequential gene modification in Pseudomonas using PCR product flanked by short homology regions. BMC Microbiol. 10, 209 (2010).
Lesic, B. & Rahme, L.G. Use of the lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol. Biol. 9, 20 (2008).
Starkey, M. et al. Pseudomonas aeruginosa rugose small colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191, 3492–3503 (2009).
Fazli, M., Harrison, J.J., Gambino, M., Givskov, M. & Tolker-Nielsen, T. In-frame and unmarked gene deletions in Burkholderia cenocepacia via an allelic exchange system compatible with Gateway technology. Appl. Environ. Microbiol. 81, 3623–3630 (2015).
Barrett, A.R. et al. Genetic tools for allelic replacement in Burkholderia species. Appl. Environ. Microbiol. 74, 4498–4508 (2008).
Lopez, C.M., Rholl, D.A., Trunck, L.A. & Schweizer, H.P. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl. Environ. Microbiol. 75, 6496–6503 (2009).
Reyrat, J.M., Pelicic, V., Gicquel, B. & Rappuoli, R. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 66, 4011–4017 (1998).
Schweizer, H. Bacterial genetics: past achievements, present state of the field, and future challenges. Biotechniques 44, 633–634, 636–641 (2008).
Lee, S.A. et al. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 112, 5189–5194 (2015).
Schweizer, H.P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97, 109–112 (1991).
Choi, K.-H. et al. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2, 443–448 (2005).
Hoang, T.T., Kutchma, A.J., Becher, A. & Schweizer, H.P. Integration proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72 (2000).
Green, M.R. & Sambrook, J. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2012).
Ausubel, F.M. et al. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology 5th edn., (John Wiley & Sons, 2002).
Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Xu, L. et al. Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between the two kingdoms. Mol. Biol. Evol. 23, 1107–1108 (2006).
Penterman, J. et al. Rapid evolution of culture-impaired bacteria during adaptation to biofilm growth. Cell Rep. 6, 293–300 (2014).
Horton, R.M., Hunt, H.D., Ho, S.N., Pullen, J.K. & Pease, L.R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 (1989).
Baraquet, C., Murakami, K., Parsek, M.R. & Harwood, C.S. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40, 7207–7218 (2012).
Ma, L. et al. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5, e1000354 (2009).
Güvener, Z.T., Tifrea, D.F. & Harwood, C.S. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol. Microbiol. 61, 106–118 (2006).
Huangyutitham, V., Guvener, Z.T. & Harwood, C.S. Subcellular clustering of the phosphorylated WspR response regulator protein stimulates its diguanylate cyclase activity. mBio 4, e00242 (2013).
Guvener, Z.T. & Harwood, C.S. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66, 1459–1473 (2007).
Heckman, K.L. & Pease, L.R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932 (2007).
van Ditmarsch, D. et al. Convergent evolution of hyperswarming leads to impaired biofilm formation in pathogenic bacteria. Cell Rep. 4, 697–708 (2013).
Rual, J.-F. et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 14, 2128–2135 (2004).
Bernard, P., Gabant, P., Bahassi, E.M. & Couturier, M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene 148, 71–74 (1994).
Vogel, H.J. & Bonner, D.M. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218, 97–106 (1956).
Blount, Z.D., Barrick, J.E., Davidson, C.J. & Lenski, R.E. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489, 513–518 (2012).
Thoma, S. & Schobert, M. An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol. Lett. 294, 127–132 (2009).
Grundemann, D. & Schomig, E. Protection of DNA during preparative agarose gel electrophoresis against damage induced by ultraviolet light. Biotechniques 21, 898–903 (1996).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Chenna, R. et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 (2003).
Leighton, T.L., Buensuceso, R., Howell, P.L. & Burrows, L.L. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ. Microbiol. doi: 10.1111/1462-2920.12849 (2015).
Burrows, L.L. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520 (2012).
Darzins, A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol. Microbiol. 11, 137–153 (1994).
Harrison, J.J. et al. Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat. Protoc. 5, 1236–1254 (2010).
Luria, S.E. & Delbruck, M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511 (1943).
Hickman, J.W., Tifrea, D.F. & Harwood, C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 102, 14422–14427 (2005).
Rietsch, A., Vallet-Gely, I., Dove, S.L. & Mekalanos, J.J. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 102, 8006–8011 (2005).
Hartley, J.L., Temple, G.F. & Brasch, M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 10, 1788–1795 (2000).
Katzen, F. Gateway recombinational cloning: a biological operating system. Expert Opin. Drug Discov. 2, 571–589 (2007).
Liang, X., Peng, L., Baek, C.H. & Katzen, F. Single step BP/LR combined Gateway reactions. Biotechniques 55, 265–268 (2013).
Stover, C.K. et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964 (2000).
Kibbe, W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35, W43–W46 (2007).
Acknowledgements
The authors thank M.F. Hynes, M. Wilton, D. van Ditmarsch, J.D. Rich and G. Winsor for useful discussions and feedback on this manuscript. J.J.H. is supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a Canada Research Chair (Tier II) from the Canadian Institutes for Health Research (CIHR). Infrastructure has been provided to J.J.H. through support from the Canada Foundation for Innovation (CFI). M.R.P. was supported by a National Institute for Allergy and Infectious Disease Grant (2R01AI077628-05A1).
Author information
Authors and Affiliations
Contributions
L.R.H. and J.J.H. wrote the manuscript; L.R.H., B.R.B., H.A., Y.I., T.E.R., M.E.L., J.J.Y., B.S.T., K.M.S., R.J.S., P.L.H., P.K.S., T.T.-N., M.R.P., H.P.S. and J.J.H. designed, tested and/or troubleshot the protocol; L.R.H., H.A., T.E.R., M.E.L., C.L. and J.J.H. created strains and plasmids; T.E.R., M.E.L., C.L. and J.J.H. generated data presented in ANTICIPATED RESULTS and Supplementary Information. All authors revised and proofread the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Irgasan (Triclosan®) and chlroamphenicol select against Pseudomonas aeruginosa, which may lead to protocol failure.
Bacteria from replicate biparental bacterial matings were suspended in 1.0 ml phosphate buffered saline (Step 36A.ix), then divided into three equal aliquots and plated on Vogel-Bonner minimal medium (VBMM) agar, or lysogeny broth (LB) agar containing 25 μg/ml irgasan (Irg) or 10 μg/ml chloramphenicol (Cm). All agar contained 60 μg/ml gentamicin (Gm) to select for merodiploids. Solid lines and error bars represent the mean number and standard deviation of P. aeruginosa merodiploids recovered after conjugation. By comparison to nutritional selection on VBMM agar, the use of Irg and Cm in selective LB agar media reduced the number of merodiploids recovered during selection.
Supplementary Figure 2 Step-by-step outcomes for building an allelic exchange vector and genome engineering.
This example focuses on building a deletion allele for pelF and creating a knockout in P. aeruginosa PAO1. (A) Vector selection and primer design (Step 1, see Supplementary Tutorial). (B) Results of PCR to synthesize mutant allele (Steps 7 and 12). Each lane for a target amplicon corresponds to a different temperature on a thermal gradient (from left to right: 70.8, 68.7, 65.6 and 61.7 °C). (C) Screening for insertion of the ΔpelF allele into pDONRPEX18Gm (Step 27). Each lane corresponds to a different E. coli clone isolated at Step 20. The band at 1253 bp corresponds to the anticipated size of the cloned ΔpelF allele (VC, vector control with no insert). (D) PCR to detect the ΔpelF mutation in the PAO1 cell line (Step 44 and 45). Each lane corresponds to a different P. aeruginosa colony isolated from NSLB + 15% sucrose agar (Step 38). The wild type pelF and ΔpelF alleles are anticipated to produce bands at 2476 and 1009 bp, respectively. This PCR step is used to identify the desired mutants, which are picked off of agar plates made at Step 39. (E) Growth of the chosen colonies on selective media (Step 39). Arrows indicate the P. aeruginosa PAO1 ΔpelF mutants that were selected based on PCR results in the previous panel and subsequently verified by sequencing (Step 47). Due to cloning efficiency in E. coli (Steps 13-19) and the theoretical 50% probability that a second crossover will fix a mutant allele in the chromosome, 4 colonies have been screened in Panels C, D and E.
Supplementary Figure 3 Proof-of-principle: A genetic dissection of biofilm matrix usage in P. aeruginosa PAO1.
Bacteria were grown in 3 ml LB overnight in 16 mm culture tubes at 37 °C at 250 rpm. The spent medium and planktonic cells were removed by aspiration. The adherent biomass was stained with crystal violet and rinsed to remove residual stain and loose material. A ΔwspF strain overproduces biofilm. Biofilm production requires the extracellular polymers Pel and Psl. Strains with ΔpelF or ΔpslD mutations produce less biofilm, whereas a strain with ΔpelF and ΔpslD mutations produces no biofilm.
Supplementary Figure 4 E. coli S17.1 and SM10 strains are functionally equivalent donor strains.
Each datum point represents an independent mating with P. aeruginosa PAO1 and either E. coli S17.1 or SM10 bearing one of four different deletion alleles (in pDONRPEX18Gm). Solid lines and error bars represent the mean and standard deviation of P. aeruginosa merodiploids recovered after conjugation with each of the E. coli donor strains, respectively.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tutorial, Supplementary Tables 1–3 (PDF 2359 kb)
Rights and permissions
About this article
Cite this article
Hmelo, L., Borlee, B., Almblad, H. et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10, 1820–1841 (2015). https://doi.org/10.1038/nprot.2015.115
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.115
This article is cited by
-
Biosynthesis pathways of expanding carbon chains for producing advanced biofuels
Biotechnology for Biofuels and Bioproducts (2023)
-
A suite of modular, all-synthetic suicide vectors for allelic exchange mutagenesis in multidrug resistant Acinetobacter strains
BMC Microbiology (2023)
-
The ColR/S two-component system is a conserved determinant of host association across Pseudomonas species
The ISME Journal (2023)
-
Bacterial c-di-GMP has a key role in establishing host–microbe symbiosis
Nature Microbiology (2023)
-
Precise genome engineering in Pseudomonas using phage-encoded homologous recombination and the Cascade–Cas3 system
Nature Protocols (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.