Abstract
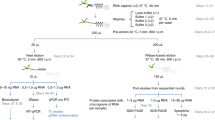
Because RNA-protein interactions have a central role in a wide array of biological processes, methods that enable a quantitative assessment of these interactions in a high-throughput manner are in great demand. Recently, we developed the high-throughput sequencing–RNA affinity profiling (HiTS-RAP) assay that couples sequencing on an Illumina GAIIx genome analyzer with the quantitative assessment of protein-RNA interactions. This assay is able to analyze interactions between one or possibly several proteins with millions of different RNAs in a single experiment. We have successfully used HiTS-RAP to analyze interactions of the EGFP and negative elongation factor subunit E (NELF-E) proteins with their corresponding canonical and mutant RNA aptamers. Here we provide a detailed protocol for HiTS-RAP that can be completed in about a month (8 d hands-on time). This includes the preparation and testing of recombinant proteins and DNA templates, clustering DNA templates on a flowcell, HiTS and protein binding with a GAIIx instrument, and finally data analysis. We also highlight aspects of HiTS-RAP that can be further improved and points of comparison between HiTS-RAP and two other recently developed methods, quantitative analysis of RNA on a massively parallel array (RNA-MaP) and RNA Bind-n-Seq (RBNS), for quantitative analysis of RNA-protein interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Amaral, P.P., Dinger, M.E., Mercer, T.R. & Mattick, J.S. The eukaryotic genome as an RNA machine. Science 319, 1787–1789 (2008).
Kruger, K. et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157 (1982).
Prody, G.A., Bakos, J.T., Buzayan, J.M., Schneider, I.R. & Bruening, G. Autolytic processing of dimeric plant virus satellite RNA. Science 231, 1577–1580 (1986).
Staley, J.P. & Woolford, J.L. Jr. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 21, 109–118 (2009).
Baltz, A.G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Furey, T.S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 13, 840–852 (2012).
Konig, J., Zarnack, K., Luscombe, N.M. & Ule, J. Protein-RNA interactions: new genomic technologies and perspectives. Nat. Rev. Genet. 13, 77–83 (2011).
Ray, D. et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat. Biotechnol. 27, 667–670 (2009).
Ray, D. et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013).
Berger, M.F. et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 24, 1429–1435 (2006).
Cho, M. et al. Quantitative selection and parallel characterization of aptamers. Proc. Natl. Acad. Sci. USA 110, 18460–18465 (2013).
Nutiu, R. et al. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 29, 659–664 (2011).
Tome, J.M. et al. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat. Methods 11, 683–688 (2014).
Mohanty, B.K., Sahoo, T. & Bastia, D. The relationship between sequence-specific termination of DNA replication and transcription. EMBO J. 15, 2530–2539 (1996).
Buenrostro, J.D. et al. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat. Biotechnol. 32, 562–568 (2014).
Shaner, N.C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004).
Ellinger, T. & Ehricht, R. Single-step purification of T7 RNA polymerase with a 6-histidine tag. Biotechniques 24, 718–720 (1998).
Guajardo, R. & Sousa, R. Characterization of the effects of Escherichia coli replication terminator protein (Tus) on transcription reveals dynamic nature of the tus block to transcription complex progression. Nucleic Acids Res. 27, 2814–2824 (1999).
Mohanty, B.K., Sahoo, T. & Bastia, D. Mechanistic studies on the impact of transcription on sequence-specific termination of DNA replication and vice versa. J. Biol. Chem. 273, 3051–3059 (1998).
Lambert, N. et al. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell 54, 887–900 (2014).
Fujita, K. & Silver, J. Surprising lability of biotin-streptavidin bond during transcription of biotinylated DNA bound to paramagnetic streptavidin beads. Biotechniques 14, 608–617 (1993).
Johansson, H.E. et al. A thermodynamic analysis of the sequence-specific binding of RNA by bacteriophage MS2 coat protein. Proc. Natl. Acad. Sci. USA 95, 9244–9249 (1998).
Gravina, M.T., Lin, J.H. & Levine, S.S. Lane-by-lane sequencing using Illumina's Genome Analyzer II. Biotechniques 54, 265–269 (2013).
Dean, K.M. & Palmer, A.E. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat. Chem. Biol. 10, 512–523 (2014).
Shi, X. et al. Quantitative fluorescence labeling of aldehyde-tagged proteins for single-molecule imaging. Nat. Methods 9, 499–503 (2012).
McCullum, E.O., Williams, B.A., Zhang, J. & Chaput, J.C. Random mutagenesis by error-prone PCR. Methods Mol. Biol. 634, 103–109 (2010).
Pluthero, F.G. Rapid purification of high-activity Taq DNA polymerase. Nucleic Acids Res. 21, 4850–4851 (1993).
Wang, Y. et al. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 32, 1197–1207 (2004).
Sambrook, J. & Russell, D.W. Preparation of denaturing polyacrylamide gels. Cold Spring Harb. Protoc. 10.1101/pdb.prot3793 (2006).
Hellman, L.M. & Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2, 1849–1861 (2007).
Lee, E.H., Kornberg, A., Hidaka, M., Kobayashi, T. & Horiuchi, T. Escherichia coli replication termination protein impedes the action of helicases. Proc. Natl. Acad. Sci. USA 86, 9104–9108 (1989).
Kornberg, R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA 104, 12955–12961 (2007).
Steitz, T.A. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr. Opin. Struct. Biol. 19, 683–690 (2009).
Lee, E.H. & Kornberg, A. Features of replication fork blockage by the Escherichia coli terminus-binding protein. J. Biol. Chem. 267, 8778–8784 (1992).
Katsamba, P.S., Park, S. & Laird-Offringa, I.A. Kinetic studies of RNA-protein interactions using surface plasmon resonance. Methods 26, 95–104 (2002).
Hall, K.B. & Kranz, J.K. Nitrocellulose filter binding for determination of dissociation constants. Methods Mol. Biol. 118, 105–114 (1999).
Rio, D.C. Filter-binding assay for analysis of RNA-protein interactions. Cold Spring Harb. Protoc. 2012, 1078–1081 (2012).
Moore, D.D. Commonly used reagents and equipment. Curr. Protoc. Mol. Biol. A.2.1–A.2.8 (2001).
Sambrook, J. & Russell, D.W. Molecular Cloning: A Laboratory Manual 3rd edn. (Cold Spring Harbor Laboratory Press, 2001).
Sambrook, J. & Russell, D.W. SDS-polyacrylamide gel electrophoresis of proteins. Cold Spring Harb. Protoc. 10.1101/pdb.prot4540 (2006).
Fairbanks, G., Steck, T.L. & Wallach, D.F. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 10, 2606–2617 (1971).
Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Pagano, J.M. et al. Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet. 10, e1004090 (2014).
Shui, B. et al. RNA aptamers that functionally interact with green fluorescent protein and its derivatives. Nucleic Acids Res. 40, e39 (2012).
Acknowledgements
We thank J.M. Pagano (Cornell University) for providing NELF-E aptamer and protein constructs, and performing NELF-E EMSA experiments; C.B. Burge (Massachusetts Institute of Technology) for the development of the seminal HiTS-FLIP assay and the scripts used for extraction of protein binding data; B. Mohanty (Medical University of South Carolina) for providing vectors containing the tus gene; K. Szeto and D. Shalloway (Cornell University) for advice on data analysis; W. Zipfel and A. Singh (Cornell University) for help in understanding the optics of the GAIIx; A. Rizzi, C.T. Waters and H. Kwak (Cornell University) for bioinformatics advice; the Cornell Sequencing Core Facility for help in running the GAIIx; and H. Craighead (Cornell University) and the members of the Lis laboratory for helpful discussions on experimental design and the manuscript. This work was supported by US National Institutes of Health grants GM090320 and DA030329 to J.T.L.
Author information
Authors and Affiliations
Contributions
A.O. and J.T.L. conceived of HiTS-RAP. A.O., J.M.T., D.G., G.P.S. and J.T.L. designed the HiTS-RAP protocol. D.G. and G.P.S. supplied sequencing reagents and equipment, as well as technical information. A.O. and J.M.T. performed the experiments. J.M.T. wrote the .xml recipe. J.M.T. and R.C.F. wrote the analysis pipeline. A.O., J.M.T. and J.T.L. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
D.G. and G.P.S. are employees of Illumina. All other authors declare no competing financial interests.
Supplementary information
Supplementary Tutorial and Software
Supplementary Tutorial 1 and Supplementary Software 1 (PDF 1179 kb)
Supplementary Software 2
extract_intensity_cif.pl (TXT 6 kb)
Supplementary Software 3
ExtractIntensitiesFromCIF_max.py (TXT 1 kb)
Supplementary Software 4
ExtractIntensitiesFromCIF_T.py (TXT 1 kb)
Supplementary Software 5
HiTS_RAP_Pipeline_Final.py (TXT 19 kb)
Rights and permissions
About this article
Cite this article
Ozer, A., Tome, J., Friedman, R. et al. Quantitative assessment of RNA-protein interactions with high-throughput sequencing–RNA affinity profiling. Nat Protoc 10, 1212–1233 (2015). https://doi.org/10.1038/nprot.2015.074
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.074
This article is cited by
-
Massively parallel profiling of RNA-targeting CRISPR-Cas13d
Nature Communications (2024)
-
Simple synthesis of massively parallel RNA microarrays via enzymatic conversion from DNA microarrays
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.