Abstract
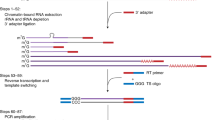
Structure-seq is a high-throughput and quantitative method that provides genome-wide information on RNA structure at single-nucleotide resolution. Structure-seq can be performed both in vivo and in vitro to study RNA structure-function relationships, RNA regulation of gene expression and RNA processing. Structure-seq can be carried out by an experienced molecular biologist with a basic understanding of bioinformatics. Structure-seq begins with chemical RNA structure probing under single-hit kinetics conditions. Certain chemical modifications, e.g., methylation of the Watson-Crick face of unpaired adenine and cytosine residues by dimethyl sulfate, result in a stop in reverse transcription. Modified RNA is then subjected to reverse transcription using random hexamer primers, which minimizes 3′ end bias; reverse transcription proceeds until it is blocked by a chemically modified residue. Resultant cDNAs are amplified by adapter-based PCR and subjected to high-throughput sequencing, subsequently allowing retrieval of the structural information on a genome-wide scale. In contrast to classical methods that provide information only on individual transcripts, a single structure-seq experiment provides information on tens of thousands of RNA structures in ∼1 month. Although the procedure described here is for Arabidopsis thaliana seedlings in vivo, structure-seq is widely applicable, thereby opening new avenues to explore RNA structure–function relationships in living organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Senecoff, J.F. & Meagher, R.B. In vivo analysis of plant RNA structure: soybean 18S ribosomal and ribulose-1,5-bisphosphate carboxylase small subunit RNAs. Plant Mol. Biol. 18, 219–234 (1992).
Tijerina, P., Mohr, S. & Russell, R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2, 2608–2623 (2007).
Wells, S.E., Hughes, J.M., Igel, A.H. & Ares, M. Jr. Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 318, 479–493 (2000).
Zaug, A.J. & Cech, T.R. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1, 363–374 (1995).
Kwok, C.K., Ding, Y., Tang, Y., Assmann, S.M. & Bevilacqua, P.C. Determination of in vivo RNA structure in low-abundance transcripts. Nat. Commun. 4, 2971 (2013).
Incarnato, D., Neri, F., Anselmi, F. & Oliviero, S. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 15, 491 (2014).
Kertesz, M. et al. Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107 (2010).
Li, F. et al. Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell 24, 4346–4359 (2012).
Underwood, J.G. et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods 7, 995–1001 (2010).
Wan, Y. et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505, 706–709 (2014).
Ding, Y. et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700 (2014).
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M. & Weissman, J.S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2014).
Talkish, J., May, G., Lin, Y., Woolford, J.L. Jr. & McManus, C.J. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA 20, 713–720 (2014).
Ehresmann, C. et al. Probing the structure of RNAs in solution. Nucleic Acids Res. 15, 9109–9128 (1987).
Spitale, R.C. et al. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 9, 18–20 (2013).
Hafner, M. et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 17, 1697–1712 (2011).
Blondal, T. et al. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res. 33, 135–142 (2005).
Aviran, S. & Pachter, L. Rational experiment design for sequencing-based RNA structure mapping. RNA 20, 1864–1877 (2014).
Siegfried, N.A., Busan, S., Rice, G.M., Nelson, J.A. & Weeks, K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods. 11, 959–965 (2014).
Tang, Y. et al. StructureFold: genome-wide RNA secondary structure mapping and reconstruction in vivo. Bioinformatics doi:10.1093/bioinformatics/btv213 (16 April 2015).
Landfors, M., Philip, P., Ryden, P. & Stenberg, P. Normalization of high dimensional genomics data where the distribution of the altered variables is skewed. PLoS ONE 6, e27942 (2011).
Deigan, K.E., Li, T.W., Mathews, D.H. & Weeks, K.M. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA 106, 97–102 (2009).
Low, J.T. & Weeks, K.M. SHAPE-directed RNA secondary structure prediction. Methods 52, 150–158 (2010).
Reuter, J.S. & Mathews, D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129 (2010).
Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011).
Goecks, J., Nekrutenko, A., Taylor, J. & Galaxy, T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 (2010).
Misra, V.K. & Draper, D.E. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 317, 507–521 (2002).
Wan, Y. et al. Genome-wide measurement of RNA folding energies. Mol. Cell 48, 169–181 (2012).
Head, S.R. et al. Library construction for next-generation sequencing: overviews and challenges. Biotechniques 56, 61–77 (2014).
Kwok, C.K., Tang, Y., Assmann, S.M. & Bevilacqua, P.C. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 40, 221–232 (2015).
Lucks, J.B. et al. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. USA 108, 11063–11068 (2011).
Lou, D.I. et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc. Natl. Acad. Sci. USA 110, 19872–19877 (2013).
Lareau, L.F., Hite, D.H., Hogan, G.J. & Brown, P.O. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife 3, e01257 (2014).
Lamm, A.T., Stadler, M.R., Zhang, H., Gent, J.I. & Fire, A.Z. Multimodal RNA-seq using single-strand, double-strand, and CircLigase-based capture yields a refined and extended description of the C. elegans transcriptome. Genome Res. 21, 265–275 (2011).
Ingolia, N.T., Brar, G.A., Rouskin, S., McGeachy, A.M. & Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
Gansauge, M.T. & Meyer, M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 8, 737–748 (2013).
Spitale, R.C. et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490 (2015).
Lawley, P.D. & Brookes, P. Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem. J. 89, 127–138 (1963).
Harris, K.A. Jr., Crothers, D.M. & Ullu, E. In vivo structural analysis of spliced leader RNAs in Trypanosoma brucei and Leptomonas collosoma: a flexible structure that is independent of cap4 methylations. RNA 1, 351–362 (1995).
Cordero, P., Kladwang, W., VanLang, C.C. & Das, R. Quantitative dimethyl sulfate mapping for automated RNA secondary structure inference. Biochemistry 51, 7037–7039 (2012).
Hajdin, C.E. et al. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. USA 110, 5498–5503 (2013).
McGinnis, J.L., Dunkle, J.A., Cate, J.H. & Weeks, K.M. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 134, 6617–6624 (2012).
Weeks, K.M. & Mauger, D.M. Exploring RNA structural codes with SHAPE chemistry. Acc. Chem. Res. 44, 1280–1291 (2011).
Wilkinson, K.A., Merino, E.J. & Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 (2006).
Wan, Y., Qu, K., Ouyang, Z. & Chang, H.Y. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 8, 849–869 (2013).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Kurtz, S., Narechania, A., Stein, J.C. & Ware, D. A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes. BMC Genomics 9, 517 (2008).
Acknowledgements
This protocol was developed under support from the Human Frontier Science Program (HFSP) grant RGP0002/2009-C, the Penn State Eberly College of Science and a Penn State Huck Huck Innovative & Transformational Seed (HITS) grant to P.C.B. and S.M.A., with additional support from NSF-IOS-1339282. We thank Y. Zhang for statistical advice; F. Pugh, Y. Li, A. Chan and K. Yen for help with Illumina sequencing; M. Axtell for helpful discussions; and P. Raghavan for access to the CyberSTAR server funded by the National Science Foundation through grant OCI-0821527. We thank L. Ritchey and Z. Su for helpful comments on the manuscript.
Author information
Authors and Affiliations
Contributions
All authors developed the protocol, designed and interpreted the experiments and wrote the paper; Y.D. and C.K.K. performed the experiments; and Y.D., C.K.K. and Y.T. analyzed the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Discussion (PDF 137 kb)
Rights and permissions
About this article
Cite this article
Ding, Y., Kwok, C., Tang, Y. et al. Genome-wide profiling of in vivo RNA structure at single-nucleotide resolution using structure-seq. Nat Protoc 10, 1050–1066 (2015). https://doi.org/10.1038/nprot.2015.064
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.064
This article is cited by
-
Genome-wide probing of eukaryotic nascent RNA structure elucidates cotranscriptional folding and its antimutagenic effect
Nature Communications (2023)
-
In vivo secondary structural analysis of Influenza A virus genomic RNA
Cellular and Molecular Life Sciences (2023)
-
Enhanced transcriptome-wide RNA G-quadruplex sequencing for low RNA input samples with rG4-seq 2.0
BMC Biology (2022)
-
RNA G-quadruplex structure contributes to cold adaptation in plants
Nature Communications (2022)
-
In vivo nuclear RNA structurome reveals RNA-structure regulation of mRNA processing in plants
Genome Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.