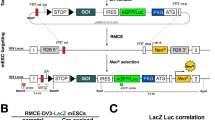
Abstract
Loss-of-function (LOF) experiments targeting multiple genes during tumorigenesis can be implemented using pooled shRNA libraries. RNAi screens in animal models rely on the use of multiple shRNAs to simultaneously disrupt gene function, as well as to serve as barcodes for cell fate outcomes during tumorigenesis. Here we provide a protocol for performing RNAi screens in orthotopic mouse tumor models, referring to glioma and lung adenocarcinoma as specific examples. The protocol aims to provide guidelines for applying RNAi to a diverse spectrum of solid tumors and to highlight crucial considerations when designing and performing these studies. It covers shRNA library assembly and packaging into lentiviral particles, and transduction into tumor-initiating cells (TICs), followed by in vivo transplantation, tumor DNA recovery, sequencing and analysis. Depending on the target genes and tumor model, tumor suppressors and oncogenes can be identified or biological pathways can be dissected in 6–9 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Uren, A.G., Kool, J., Berns, A. & van Lohuizen, M. Retroviral insertional mutagenesis: past, present and future. Oncogene 24, 7656–7672 (2005).
Brummelkamp, T.R., Bernards, R. & Agami, R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2, 243–247 (2002).
Zender, L. et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 (2008).
Bric, A. et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell 16, 324–335 (2009).
Meacham, C.E., Ho, E.E., Dubrovsky, E., Gertler, F.B. & Hemann, M.T. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat. Genet. 41, 1133–1137 (2009).
Gargiulo, G. et al. In vivo RNAi screen for BMI1 targets identifies TGF-β/BMP-ER stress pathways as key regulators of neural- and malignant glioma–stem cell homeostasis. Cancer Cell 23, 660–676 (2013).
Xue, W. et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc. Natl. Acad. Sci. USA 109, 8212–8217 (2012).
Iorns, E. et al. Whole-genome in vivo RNAi screening identifies the leukemia inhibitory factor receptor as a novel breast tumor suppressor. Breast Cancer Res. Treat. 135, 79–91 (2012).
Miller, P.G. et al. In vivo RNAi screening identifies a leukemia-specific dependence on integrin-β3 signaling. Cancer Cell 24, 45–58 (2013).
Braun, C.J. & Hemann, M.T. Unraveling tumor suppressor networks with in vivo RNAi. Cell Stem Cell 12, 639–641 (2013).
O'Brien, C.A., Kreso, A. & Jamieson, C.H.M. Cancer stem cells and self-renewal. Clin. Cancer Res. 16, 3113–3120 (2010).
Magee, J.A., Piskounova, E. & Morrison, S.J. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell 21, 283–296 (2012).
Beronja, S. et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 501, 185–190 (2013).
Root, D.E., Hacohen, N., Hahn, W.C., Lander, E.S. & Sabatini, D.M. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods 3, 715–719 (2006).
Huang, S. et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell 15, 328–340 (2009).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Wuestefeld, T. et al. A direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell 153, 389–401 (2013).
Wang, T., Wei, J.J., Sabatini, D.M. & Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Du, J. et al. In vivo RNAi screen reveals neddylation genes as novel regulators of Hedgehog signaling. PLoS ONE 6, e24168 (2011).
Chen, T. et al. An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration. Nature 485, 104–108 (2012).
Kaelin, W.G. Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science 337, 421–422 (2012).
Wilson, J.L., Hemann, M.T., Fraenkel, E. & Lauffenburger, D.A. Integrated network analyses for functional genomic studies in cancer. Semin. Cancer Biol. 23, 213–218 (2013).
Lee, D.-F. et al. Combining competition assays with genetic complementation strategies to dissect mouse embryonic stem cell self-renewal and pluripotency. Nat. Protoc. 7, 729–748 (2012).
Bruggeman, S.W.M. et al. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell 12, 328–341 (2007).
Ellis, B.L., Potts, P.R. & Porteus, M.H. Creating higher titer lentivirus with caffeine. Hum. Gene Ther. 22, 93–100 (2011).
Kutner, R.H., Zhang, X.-Y. & Reiser, J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 4, 495–505 (2009).
Baumann, B.C., Dorsey, J.F., Benci, J.L., Joh, D.Y. & Kao, G.D. Stereotactic intracranial implantation and in vivo bioluminescent imaging of tumor xenografts in a mouse model system of glioblastoma multiforme. J. Vis. Exp. 67, e4089 (2012).
Montini, E. et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 119, 964–975 (2009).
Zuber, J. et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 29, 79–83 (2011).
Fellmann, C. et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013).
Wolf, J. et al. An in vivo RNAi screen identifies SALL1 as a tumor suppressor in human breast cancer with a role in CDH1 regulation. Oncogene 33, 4273–4278 (2013).
Acknowledgements
This protocol is the in vivo evolution of the in vitro RNAi screen set up by the Agami, Beijersbergen and Bernards laboratories, which are acknowledged. We are thankful to the NKI Genomics Core (R. Kerkhoven and I. de Rink), as well as to Microscopy, Screening (B. Morris), FACS and Animal Facilities for assistance with experiments, and to the following persons for expert advice: M. Amendola (virus packaging and titration), P. Possik and K. Greig (amplification and barcode-tagging of shRNA libraries) and J. Jaspers (tumor cell isolation). We are grateful to K. Sutherland and A. Berns (both from the Netherlands Cancer Institute) for KrasG12D/+;Trp53−/− cells and B. Siteur for excellent assistance with animal experiments, as well as to W. Akhtar, C. Biancotto, A. Gruszka and E. Guccione for critical input on this manuscript. This work was supported by the European Commission project (Marie Curie) FCMOG (FP7-PEOPLE-2009-IEF 251938) and Fondazione Lorini to G.G., by the Koningin Wilhelmina Fonds (KWF) project NKI2013-6030 to G.G. and M.v.L., and by the Netherlands Genomics Initiative to M.v.L.
Author information
Authors and Affiliations
Contributions
G.G. designed and developed the procedure and wrote the manuscript. M.S., D.H. and M.C. helped with experiments and data analysis. M.v.L. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
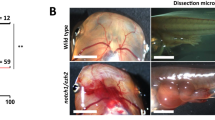
Supplementary Figure 1 Example of NSCLC cell grafting efficiency.
Mouse lung adenocarcinoma cells with a KrasG12D;Trp53-/- were modified with a library containing both (candidate) TSG and oncogenes and luciferase emission was acquired immediately after tail vein injection (∼30 min), 24 hours and 7 days later. a) Luciferase emission for each mouse back and belly at day 0, 1 and 7. Statistics used one-way ANOVA and Dunn test for multiple comparisons. b) The IVIS scan for two independent animals from (a) is shown at the indicated days.
All animal experiments were conducted in compliance with the European Union guidelines for the use and care of laboratory animals and were reviewed and approved by the animal ethics committee (DEC) of the Netherlands Cancer Institute.
Supplementary Figure 2 Input tables.
Example of table formatting for Box 4 and Step 49.
Supplementary Figure 3 Example of hairpin Sanger sequencing result as validation for MOI<1 infection.
Brain tumour generated with a pool of shRNA was dissociated and plated at clonal density to isolate clonal cell populations (glioma-spheres). Multiple single spheres (>20) were subjected to hairpin library amplification followed by Sanger sequencing. Successful base calling during shRNA sequencing indicates the presence of one single hairpin per cell is shown as an example (as in 6). Reproduced from ref. 6, © 2013, with permission from Elsevier.
Supplementary Figure 4 Example of correlation between two in vivo dropout screen biological replicas.
Each data point represents one normalized hairpin enrichment/depletion value in each experimental group (recipient animals #1-#2-#3-#4-#5 are correlated with #6-#7-#8-#9-#10). Data were processed as in step 48, linear regression was performed as in step 55, and axes variation is -15 to 10, lower left quadrant include dropout hairpins in KrasG12V/+;Trp53-/- cells. All animal experiments were conducted in compliance with the European Union guidelines for the use and care of laboratory animals and were reviewed and approved by the animal ethics committee (DEC) of the Netherlands Cancer Institute
Supplementary Figure 5 Example of experimental variation between hairpins.
For two different “Input” cell types and libraries, we have sequenced using a depth of ∼5,000 fold the library size and the actual reads count (Log2 transformed) is shown. These results reflect the actual variation introduced by experimental steps 1–30.
Supplementary information
Supplementary Figures 1–5
Supplementary Figures 1–5 (PDF 671 kb)
Supplementary Software: Perl script file example.
Contains the set of instructions to perform annotation of the sequencing. It should be copied into a text file and named “harpin_fast.pl” or a different name should be indicated in the perl script. Moreover, we indicated the name “barcode.txt” in case of multiplexed runs: this file is not required for the perl script to function, but it should be renamed in perl script if a different name is used for that instruction. (TXT 2 kb)
Supplementary Table 1: Example of input annotation table.
This file contains the relevant information to run Supplementary Data 1, and can be used as a template for customized hairpin libraries. It should be copied into a text file and named “Perl harpin_fast.pl annotation_file.txt”. (TXT 101 kb)
Rights and permissions
About this article
Cite this article
Gargiulo, G., Serresi, M., Cesaroni, M. et al. In vivo shRNA screens in solid tumors. Nat Protoc 9, 2880–2902 (2014). https://doi.org/10.1038/nprot.2014.185
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.185
This article is cited by
-
Stiffer-Matrix-Induced PGC-1α Upregulation Enhanced Mitochondrial Biogenesis and Oxidative Stress Resistance in Non-small Cell Lung Cancer
Cellular and Molecular Bioengineering (2023)
-
Antibody-coupled siRNA as an efficient method for in vivo mRNA knockdown
Nature Protocols (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.