Abstract
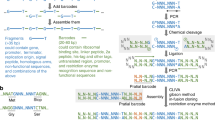
Recombination-based DNA construction methods, such as Gibson assembly, have made it possible to easily and simultaneously assemble multiple DNA parts, and they hold promise for the development and optimization of metabolic pathways and functional genetic circuits. Over time, however, these pathways and circuits have become more complex, and the increasing need for standardization and insulation of genetic parts has resulted in sequence redundancies—for example, repeated terminator and insulator sequences—that complicate recombination-based assembly. We and others have recently developed DNA assembly methods, which we refer to collectively as unique nucleotide sequence (UNS)–guided assembly, in which individual DNA parts are flanked with UNSs to facilitate the ordered, recombination-based assembly of repetitive sequences. Here we present a detailed protocol for UNS-guided assembly that enables researchers to convert multiple DNA parts into sequenced, correctly assembled constructs, or into high-quality combinatorial libraries in only 2–3 d. If the DNA parts must be generated from scratch, an additional 2–5 d are necessary. This protocol requires no specialized equipment and can easily be implemented by a student with experience in basic cloning techniques.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Keasling, J.D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 3, 64–76 (2008).
Torella, J.P. et al. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc. Natl. Acad. Sci. 110, 11290–11295 (2013).
Prather, K.L.J. & Martin, C.H. De novo biosynthetic pathways: rational design of microbial chemical factories. Curr. Opin. Biotechnol. 19, 468–474 (2008).
Savage, D.F., Way, J. & Silver, P.A. Defossiling fuel: How synthetic biology can transform biofuel production. ACS Chem. Biol. 3, 13–16 (2008).
Khalil, A.S. & Collins, J.J. Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379 (2010).
Purnick, P.E.M. & Weiss, R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 10, 410–422 (2009).
Bowen, T.A., Zdunek, J.K. & Medford, J.I. Cultivating plant synthetic biology from systems biology. New Phytol. 179, 583–587 (2008).
Lienert, F., Lohmueller, J.J., Garg, A. & Silver, P.A. Synthetic biology in mammalian cells: next-generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 15, 95–107 (2014).
Ruder, W.C., Lu, T. & Collins, J.J. Synthetic biology moving into the clinic. Science 333, 1248–1252 (2011).
Weber, W. & Fussenegger, M. Emerging biomedical applications of synthetic biology. Nat. Rev. Genet. 13, 21–35 (2012).
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012).
Bugaj, L.J. & Schaffer, D.V. Bringing next-generation therapeutics to the clinic through synthetic biology. Curr. Opin. Chem. Biol. 16, 355–361 (2012).
Chen, Y.Y., Galloway, K.E. & Smolke, C.D. Synthetic biology: advancing biological frontiers by building synthetic systems. Genome Biol. 13, 240 (2012).
Agapakis, C.M. et al. Insulation of a synthetic hydrogen metabolism circuit in bacteria. J. Biol. Eng. 4, 3 (2010).
Chen, Y.J. et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 10, 659–664 (2013).
Litcofsky, K.D., Afeyan, R.B., Krom, R.J., Khalil, A.S. & Collins, J.J. Iterative plug-and-play methodology for constructing and modifying synthetic gene networks. Nat. Methods 9, 1077–1080 (2012).
Zelcbuch, L. et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 41, e98 (2013).
Esvelt, K.M. & Wang, H.H. Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 9, 641 (2013).
Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Gibson, D.G. et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319, 1215–1220 (2008).
Gibson, D.G., Smith, H.O., Hutchison, C.A., Venter, J.C. & Merryman, C. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods 7, 901–903 (2010).
Torella, J.P. et al. Rapid construction of insulated genetic circuits via synthetic sequence-guided isothermal assembly. Nucleic Acids Res. 42, 681–689 (2014).
Lienert, F. et al. Two-and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 41, 9967–9975 (2013).
Guye, P., Li, Y., Wroblewska, L., Duportet, X. & Weiss, R. Rapid, modular and reliable construction of complex mammalian gene circuits. Nucleic Acids Res. 41, e156 (2013).
Casini, A. et al. One-pot DNA construction for synthetic biology: the Modular Overlap-Directed Assembly with Linkers (MODAL) strategy. Nucleic Acids Res. 42, e7 (2014).
Ramon, A. & Smith, H.O. Single-step linker-based combinatorial assembly of promoter and gene cassettes for pathway engineering. Biotechnol. Lett. 33, 549–555 (2011).
Du, J., Yuan, Y., Si, T., Lian, J. & Zhao, H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 40, e142 (2012).
Knight, T.F. Idempotent vector design for standard assembly of biobricks MIT Artificial Intelligence Laboratory http://dspace.mit.edu/handle/1721.1/21168 (2003).
Anderson, J.C. et al. BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 4, 1 (2010).
Borujeni, A.E., Channarasappa, A.S. & Salis, H.M. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 42, 2646–2659 (2014).
Engler, C., Kandzia, R. & Marillonnet, S. A one-pot, one-step, precision cloning method with high throughput capability. PLoS ONE 3, e3647 (2008).
Engler, C., Gruetzner, R., Kandzia, R. & Marillonnet, S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4, e5553 (2009).
Quan, J. & Tian, J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4, e6441 (2009).
Quan, J. & Tian, J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 6, 242–251 (2011).
Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K. & Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989).
Xiong, A.S. et al. PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 1, 791–797 (2006).
de Kok, S. et al. Rapid and reliable DNA assembly via ligase cycling reaction (LCR). ACS Synth. Biol. 3, 97–106 (2014).
Bayer, T.S. et al. Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 131, 6508–6515 (2009).
Balibar, C.J. & Walsh, C.T. In vitro biosynthesis of violacein from L-tryptophan by the enzymes VioA-E from Chromobacterium violaceum. Biochemistry 45, 15444–15457 (2006).
Acknowledgements
The authors thank C.T. Walsh, T.A. Wencewicz, P. Malkus and J. Paulsson for plasmids and reagents, and T.J. Ford, D.C. MacKellar, A.H. Chen and H.M. Salis for helpful discussions relating to this work. This work was supported by the Advanced Research Projects Agency-Energy 'Electrofuels' Collaborative Agreement (grant no. DE-AR0000079 to P.A.S.); a National Science Foundation Graduate Research Fellowship and Herchel Smith Graduate Research Fellowship to J.P.T.; a European Molecular Biology Organization and Human Frontier Science Program Fellowship to F.L.; a German National Academic Foundation Scholarship to C.R.B.; a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship to J.-H.C.; and the Defense Advanced Research Projects Agency grant (no. 4500000572 to P.A.S.). This material is based on work supported by the National Science Foundation. Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Contributions
J.P.T. conceived the project; J.P.T., C.R.B., F.L., J.-H.C., P.A.S. and J.C.W. contributed key ideas to the development of the project; P.A.S. and J.C.W. supervised the project; and J.P.T., C.R.B., F.L. and J.-H.C. performed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Torella, J., Lienert, F., Boehm, C. et al. Unique nucleotide sequence–guided assembly of repetitive DNA parts for synthetic biology applications. Nat Protoc 9, 2075–2089 (2014). https://doi.org/10.1038/nprot.2014.145
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.145
This article is cited by
-
Silent gene clusters encode magnetic organelle biosynthesis in a non-magnetotactic phototrophic bacterium
The ISME Journal (2023)
-
Real-time bioelectronic sensing of environmental contaminants
Nature (2022)
-
Two efficient CRISPR/Cas9 systems for gene editing in soybean
Transgenic Research (2021)
-
Fine-tuning the regulation of Cas9 expression levels for efficient CRISPR-Cas9 mediated recombination in Streptomyces
Journal of Industrial Microbiology and Biotechnology (2020)
-
Metagenomic mining of regulatory elements enables programmable species-selective gene expression
Nature Methods (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.