Abstract
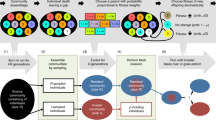
Estimation of fitness is a key step in experimental evolution studies. However, no established methods currently exist to specifically estimate how successful new alleles are in invading populations. The main reason is that most assays do not accurately reflect the randomness associated with the first stages of the invasion, when invaders are rare and extinctions are frequent. In this protocol, I describe how such experiments can be done in an effective way. By using the nematode model, Caenorhabditis elegans, a large number of invasion experiments are set up, whereby invading individuals carrying a visual marker are introduced into populations in very low numbers. The number of invaders counted in consecutive generations, together with the number of extinctions, is then used in the context of individual-based computer simulations to provide likelihood (Lk) estimates for fitness. This protocol can take up to five generations of experimental invasions and a few hours of computer processing time.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Orr, H.A. Fitness and its role in evolutionary genetics. Nat. Rev. Genet. 10, 531–539 (2009).
Hartl, D.L. & Clark, A.G. Principles of Population Genetics 3rd edn. (Sinauer Associates, 1997).
Fisher, R. On the dominance ratio. Proc. Royal Soc. Edinburgh 42, 321–341 (1922).
Haldane, J. A mathematical theory of natural and artificial selection, part V: selection and mutation. Math. Proc. Cambridge Phil. Soc. 23, 838–844 (1927).
Heffernan, J.M. & Wahl, L.M. The effects of genetic drift in experimental evolution. Theor. Popul. Biol. 62, 349–356 (2002).
Gifford, D.G., de Visser, J.A. & Wahl, L.M. Model and test in a fungus of the probability that beneficial mutations survive drift. Biol. Lett. 9, 2012.0310 (2012).
Roff, D.A. Defining fitness in evolutionary models. J. Genet. 87, 339–348 (2008).
Chelo, I.M., Nédli, J., Gordo, I. & Teotónio, H. An experimental test on the probability of extinction of new genetic variants. Nat. Commun. 4, 2417 (2013).
Fisher, R. The Genetical Theory of Natural Selection (Clarendon Press, 1930).
Stiernagle, T. Maintenance of C. elegans. in C. elegans: a Practical Approach (ed. Hope, I.A.) (Oxford University Press, 1999).
Teotónio, H., Carvalho, S., Manoel, R., Roque, M. & Chelo, I.M. Evolution of outcrossing in experimental populations of Caenorhabditis elegans. PLoS ONE 7, 1 (2012).
Chelo, I.M. & Teotónio, H. The opportunity for balancing selection in experimental populations of Caenorhabditis elegans. Evolution 67, 142–156 (2012).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Metz, J.A.J., Nisbet, R.M. & Geritz, S.A.H. How should we define 'fitness' for general ecological scenarios? Trends. Ecol. Evol. 7, 198–202 (1992).
R Development Core Team. R: A Language and Environment for Statistical Computing, http://www.R-project.org (2006).
Elena, S.F. & Lenski, R.E. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4, 457–469 (2003).
Perfeito, L., Fernandes, L., Mota, C. & Gordo, I. Adaptive mutations in bacteria: high rate and small effects. Science 317, 813–815 (2007).
Wright, S. Evolution in Mendelian populations. Genetics 16, 97–159 (1931).
Chevin, L.M. On measuring selection in experimental evolution. Biol. Lett. 7, 210–213 (2011).
Doebeli, M. & Dieckmann, U. Evolutionary branching and sympatric speciation caused by different types of ecological interactions. Am. Nat. 156, S77–S101 (2000).
Mylius, S.D. & Dieckmann, O. The resident strikes back: invader-induced switching of resident attractor. J. Theor. Biol. 211, 297–311 (2001).
Yanik, M.F., Rohde, C.B. & Pardo-Martin, C. Technologies for micromanipulating, imaging and phenotyping small invertebrates and vertebrates. Annu. Rev. Biomed. Eng. 13, 185–217 (2011).
Acknowledgements
I thank I. Gordo and H. Teotónio for advice in the development and implementation of this protocol. I also thank the reviewers for suggestions and comments, which markedly improved the manuscript. This work was only possible with the support of the Foundation for Science and Technology, Portugal (FCT Investigator program and EXPL/BIA-EFV/1211/2013) to I.M.C., and financial and technical support from Henrique Teotónio, through the Human Frontiers Science Program grant (RGP0045/2010).
Author information
Authors and Affiliations
Contributions
I.M.C. designed the protocol and wrote the simulation, the analysis script and the manuscript.
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Effect of generation number on fitness estimation.
Different sets were simulated for 2, 3, 4, 4 and 7 generations and selection coefficients of 0, 0.001, 0.01, 0.05, 0.1, 0.2, 0.3 and 0.5, 30 replicates were used. Maximum likelihood (ML) estimates and –2lnLk confidence intervals were obtained to show how generation number affects the accuracy (a), “statistical” power (see definition below) (b) and precision (c) of the estimation procedure. For each parameter combination, 100 simulations were carried out. Each invasion started with two individuals in a resident population of 1000 individuals. Data from every generation was used. ML estimation is based on 107 simulations for each of 61 selection coefficients (form –0.1 to 0.5 in steps of 0.01). (a) The mean (symbols) and standard deviation (error bars) of estimated s (ŝ) are shown. (b) The estimation of “statistical” power is provided by the proportion of times the 'true' selection coefficient is found in the –2lnLk confidence interval (mean standard deviations are shown) reveals how the number of generations affects precision. The increase in number of possible trajectories with the number of generations results in a lower power and downward bias in the estimation of selection coefficients.
Supplementary information
Supplementary Figure 1
Effect of generation number on fitness estimation. (PDF 2994 kb)
Supplementary Table 1
Observed counts from an experiment starting with two invading individuals where competitions experiments ran for five generations and scoring was obtained for generations 3 through 5. (TXT 0 kb)
Supplementary Data
A script file in the R language which gives the implementation in R code of the invasion simulation and estimation of selection coefficients described in steps 24 through 31. (TXT 5 kb)
Rights and permissions
About this article
Cite this article
Chelo, I. Experimental determination of invasive fitness in Caenorhabditis elegans. Nat Protoc 9, 1392–1400 (2014). https://doi.org/10.1038/nprot.2014.098
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.098
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.