Abstract
This protocol describes how to observe gastrulation in living mouse embryos by using light-sheet microscopy and computational tools to analyze the resulting image data at the single-cell level. We describe a series of techniques needed to image the embryos under physiological conditions, including how to hold mouse embryos without agarose embedding, how to transfer embryos without air exposure and how to construct environmental chambers for live imaging by digital scanned light-sheet microscopy (DSLM). Computational tools include manual and semiautomatic tracking programs that are developed for analyzing the large 4D data sets acquired with this system. Note that this protocol does not include details of how to build the light-sheet microscope itself. Time-lapse imaging ends within 12 h, with subsequent tracking analysis requiring 3–6 d. Other than some mouse-handling skills, this protocol requires no advanced skills or knowledge. Light-sheet microscopes are becoming more widely available, and thus the techniques outlined in this paper should be helpful for investigating mouse embryogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Change history
18 April 2014
In the version of this article initially published, Table 1 contained a number of errors: lateral resolution for two-photon was erroneously stated as ‘r divided by the square root of 2’ instead of ‘r multiplied by the square root of 2’; the illumination intensity for two-photon microscopy was given as ‘103 times E’ instead of ‘106 times E’; ‘lateral resolution’ was assigned the incorrect footnote; and reference 11 was incorrectly cited as a source for the table. An additional footnote (a) has also been added to the version that was originally published. These errors have been corrected in the HTML and PDF versions of the article.
References
Yamanaka, Y., Tamplin, O.J., Beckers, A., Gossler, A. & Rossant, J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev. Cell 13, 884–896 (2007).
Kwon, G.S., Viotti, M. & Hadjantonakis, A.K. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 15, 509–520 (2008).
Huisken, J. & Stainier, D.Y. Selective plane illumination microscopy techniques in developmental biology. Development 136, 1963–1975 (2009).
Tomer, R., Khairy, K. & Keller, P.J. Shedding light on the system: studying embryonic development with light-sheet microscopy. Curr. Opin. Genet. Dev. 21, 558–565 (2011).
Khairy, K. & Keller, P.J. Reconstructing embryonic development. Genesis 49, 488–513 (2011).
Hockendorf, B., Thumberger, T. & Wittbrodt, J. Quantitative analysis of embryogenesis: a perspective for light-sheet microscopy. Dev. Cell 23, 1111–1120 (2012).
Tomer, R., Khairy, K. & Keller, P.J. Light-sheet microscopy in cell biology. Methods Mol. Biol. 931, 123–137 (2013).
Keller, P.J. & Stelzer, E.H. Digital scanned laser light-sheet fluorescence microscopy. Cold Spring Harb. Protoc. 2010 10.1101/pdb.top78 (2010).
Keller, P.J., Schmidt, A.D., Wittbrodt, J. & Stelzer, E.H. Reconstruction of zebrafish early embryonic development by scanned light-sheet microscopy. Science 322, 1065–1069 (2008).
Keller, P.J. et al. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat. Methods 7, 637–642 (2010).
Truong, T.V., Supatto, W., Koos, D.S., Choi, J.M. & Fraser, S.E. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods 8, 757–760 (2011).
Wu, Y. et al. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108, 17708–17713 (2011).
Arrenberg, A.B., Stainier, D.Y., Baier, H. & Huisken, J. Optogenetic control of cardiac function. Science 330, 971–974 (2010).
Ichikawa, T. et al. Live imaging of whole mouse embryos during gastrulation: migration analyses of epiblast and mesodermal cells. PLoS ONE 8, e64506 (2013).
Yamamoto, M. et al. Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature 428, 387–392 (2004).
Takaoka, K., Yamamoto, M. & Hamada, H. Origin and role of distal visceral endoderm, a group of cells that determines anterior-posterior polarity of the mouse embryo. Nat. Cell Biol. 13, 743–752 (2011).
Tam, P.P., Parameswaran, M., Kinder, S.J. & Weinberger, R.P. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: the role of ingression and tissue movement during gastrulation. Development 124, 1631–1642 (1997).
Quinlan, G.A., Williams, E.A., Tan, S.S. & Tam, P.P. Neuroectodermal fate of epiblast cells in the distal region of the mouse egg cylinder: implication for body plan organization during early embryogenesis. Development 121, 87–98 (1995).
Lee, J.D. & Anderson, K.V. Morphogenesis of the node and notochord: the cellular basis for the establishment and maintenance of left-right asymmetry in the mouse. Dev. Dyn. 237, 3464–3476 (2008).
Hara, K. et al. Evidence for crucial role of hindgut expansion in directing proper migration of primordial germ cells in mouse early embryogenesis. Dev. Biol. 330, 427–439 (2009).
Saitou, M. & Yamaji, M. Germ cell specification in mice: signaling, transcription regulation, and epigenetic consequences. Reproduction 139, 931–942 (2010).
Anderson, R. et al. Mouse primordial germ cells lacking β1 integrins enter the germline but fail to migrate normally to the gonads. Development 126, 1655–1664 (1999).
Keller, R. et al. Mechanisms of convergence and extension by cell intercalation. Philos. Trans. R Soc. Lond. B Biol. Sci. 355, 897–922 (2000).
Gefen, A. Studies in Mechanobiology, Tissue Engineering and Biomaterials (Springer, 2011).
Lim, J. & Thiery, J.P. Epithelial-mesenchymal transitions: insights from development. Development 139, 3471–3486 (2012).
Tomer, R., Khairy, K., Amat, F. & Keller, P.J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–763 (2012).
Sturm, K. & Tam, P.P. Isolation and culture of whole postimplantation embryos and germ layer derivatives. Methods Enzymol. 225, 164–190 (1993).
Abe, T. et al. Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis 49, 579–590 (2011).
Thevenaz, P., Ruttimann, U.E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process 7, 27–41 (1998).
Micheva, K.D., O'Rourke, N., Busse, B. & Smith, S.J. Array tomography: high-resolution three-dimensional immunofluorescence. Cold Spring Harb. Protoc. 2010 pdb top89 (2010).
Meijering, E.H., Niessen, W.J. & Viergever, M.A. Quantitative evaluation of convolution-based methods for medical image interpolation. Med. Image Anal. 5, 111–126 (2001).
Keller, P.J. In vivo imaging of zebrafish embryogenesis. Methods 62, 268–278 (2013).
Gualda, E.J. et al. OpenSpinMicroscopy: an open-source integrated microscopy platform. Nat. Methods 10, 599–600 (2013).
Nagy, A. Manipulating The Mouse Embryo: A Laboratory Manual 3rd edn. (Cold Spring Harbor Laboratory Press, 2003).
Shea, K. & Geijsen, N. Dissection of 6.5 dpc mouse embryos. J. Vis. Exp. 160 10.3791/160 (2007).
Nonaka, S. Modification of mouse nodal flow by applying artificial flow. Methods Cell Biol. 91, 287–297 (2009).
Acknowledgements
We thank T. Fujimori for Histone H2B-GFP, H2B-mCherry and Lyn-Venus transgenic mice; H. Tao for handling of the mice; and F. Härle for DSLM software. We also thank members of the S.N. laboratory, the N. Ueno laboratory and the N. Shiina laboratory for valuable discussions, comments and technical assistance. We thank N. Papas for comments on the manuscript. This work was supported by a Grant-in-Aid for Young Scientists (A) from the Japan Society for the Promotion of Science (JSPS, 18687902); by the JSPS Fellows to T.I. (22353); by the Ministry of Education, Culture, Sports, Science and Technology (MEXT, 10J00353); by the Core Research for Evolutional Science and Technology (CREST) program; and by the Human Frontier Science Program (HFSP).
Author information
Authors and Affiliations
Contributions
T.I. developed most of the protocols described here and performed the experiments with the help of H.K.-K. under the supervision of S.N. K.N. developed the analysis tools under the supervision of A.M. P.J.K. and E.H.K.S. developed DSLM. T.I. and P.J.K. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Effect of phototoxicity on mouse embryo during gastrulation using long wavelength illumination.
H2B–mCherry transgenetic mouse at embryonic day 6.5 was used. The embryo was illuminated with 647 nm laser light in 100 optical sections per image stack, once every 5 minutes. Continuous illumination with long wavelength light for 12 hours does not cause embryo degradation. Scale bar: 20 μm.
Supplementary Figure 2 Design drawing of embryo holder.
The holder is made from a 5-mm-diameter acrylic rod. The left side of the holder is glued to the piston of a 1-ml syringe.
Supplementary Figure 3 Design drawing of gas adapter.
The adapter is made from a 2-mm-thick acrylic plate.
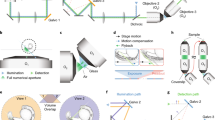
Supplementary Figure 4 Digital scanned light-sheet microscope setup and light path.
Laser beam from argon-krypton laser is selected with AOTF. A light sheet is formed using the two-axis high-speed scan head and f-theta lens. The light sheet excites fluorescent molecules in the sample. Fluorescence is detected via the detection lens, long pass filter and CCD camera.
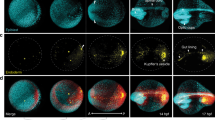
Supplementary Figure 5 Examples of optical sections from live imaging experiments with mouse embryos at embryonic day (E) 5.5 to 7.5.
Top shows nuclear stained embryo (H2B–mCherry) and bottom shows membrane stained embryo (Lyn–Venus). Scale bar = 20 μm.
Supplementary information
Supplementary Figure 1
Effect of phototoxicity on mouse embryo during gastrulation using long wavelength illumination. (PDF 155 kb)
Supplementary Figure 2
Design drawing of embryo holder. (PDF 87 kb)
Supplementary Figure 3
Design drawing of gas adapter. (PDF 82 kb)
Supplementary Figure 4
Digital scanned light-sheet microscope setup and light path. (PDF 232 kb)
Supplementary Figure 5
Examples of optical sections from live imaging experiments with mouse embryos at embryonic day (E) 5.5 to 7.5. (PDF 248 kb)
Live imaging of Histone H2B–mCherry mouse embryo at embryonic day 6.5.
The optical section shown here is located about 85 μm from the distal end of the embryo. The time interval is 5 min. (MOV 4722 kb)
Supplementary Data 1
Programs described in this protocol. The archive includes Matlab scripts for image projection and cropping, the ImageJ macro for image registration, compiled programs for manual and semi-automatic tracking and the POV-Ray script for visualizing trajectories. Descriptions of each program are provided in Table 2. (ZIP 230 kb)
Supplementary Data 2
Source code of manual and semi-automatic tracking programs. The archive includes source code for “manual_tracker.exe”, “cdots_search.exe”, “ichi_srcx.exe”, “semiauto_tracer.exe”, “cc_trace.exe” and “cc_reduce.exe”. These programs were written in C++ and C# using Visual Studio 2010 (Microsoft). (ZIP 374 kb)
Rights and permissions
About this article
Cite this article
Ichikawa, T., Nakazato, K., Keller, P. et al. Live imaging and quantitative analysis of gastrulation in mouse embryos using light-sheet microscopy and 3D tracking tools. Nat Protoc 9, 575–585 (2014). https://doi.org/10.1038/nprot.2014.035
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.035
This article is cited by
-
Light sheet fluorescence microscopy
Nature Reviews Methods Primers (2021)
-
Long-term, in toto live imaging of cardiomyocyte behaviour during mouse ventricle chamber formation at single-cell resolution
Nature Cell Biology (2020)
-
Multiscale imaging of plant development by light-sheet fluorescence microscopy
Nature Plants (2018)
-
Three-dimensional microCT imaging of murine embryonic development from immediate post-implantation to organogenesis: application for phenotyping analysis of early embryonic lethality in mutant animals
Mammalian Genome (2018)
-
Preparation of plants for developmental and cellular imaging by light-sheet microscopy
Nature Protocols (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.