Abstract
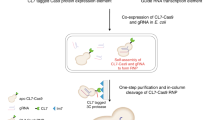
We recently invented a method to produce highly potent siRNAs in Escherichia coli, based on the serendipitous discovery that ectopic expression of p19, a plant viral siRNA-binding protein, stabilizes otherwise unstable bacterial siRNAs, which we named pro-siRNAs for prokaryotic siRNAs. We present a detailed protocol describing how to produce pro-siRNAs for efficiently knocking down any gene, beginning with the design of a pro-siRNA expression plasmid and ending with siRNA purification. This protocol uses one plasmid to co-express a recombinant His-tagged p19 protein and a long hairpin RNA containing sense and antisense sequences of the target gene. pro-siRNAs are isolated and purified using nickel beads and HPLC, using methods used to produce recombinant proteins. Once a pro-siRNA plasmid is obtained, production of purified pro-siRNAs takes a few days. The pro-siRNA technique provides a reliable and renewable source of siRNAs, and it can be implemented in any laboratory whose members are skilled in routine molecular biology techniques.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Hamilton, A.J. & Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952 (1999).
Knight, S.W. & Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269–2271 (2001).
Bohmert, K. et al. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180 (1998).
Carmell, M.A., Xuan, Z., Zhang, M.Q. & Hannon, G.J. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16, 2733–2742 (2002).
Baulcombe, D. RNA silencing in plants. Nature 431, 356–363 (2004).
Ding, S.W. & Voinnet, O. Antiviral immunity directed by small RNAs. Cell 130, 413–426 (2007).
Elbashir, S.M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Caplen, N.J., Parrish, S., Imani, F., Fire, A. & Morgan, R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98, 9742–9747 (2001).
Yang, D. et al. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 99, 9942–9947 (2002).
Myers, J.W., Jones, J.T., Meyer, T. & Ferrell, J.E. Jr. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 21, 324–328 (2003).
Kittler, R. et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat. Methods 4, 337–344 (2007).
Myers, J.W. et al. Minimizing off-target effects by using diced siRNAs for RNA interference. J. RNAi Gene Silencing 2, 181–194 (2006).
Stewart, S.A. et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003).
Zhao, H.F. et al. High-throughput screening of effective siRNAs from RNAi libraries delivered via bacterial invasion. Nat. Methods 2, 967–973 (2005).
Li, Z., Fortin, Y. & Shen, S.H. Forward and robust selection of the most potent and noncellular toxic siRNAs from RNAi libraries. Nucleic Acids Res. 37, e8 (2009).
Xiang, S., Fruehauf, J. & Li, C.J. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat. Biotechnol. 24, 697–702 (2006).
Tenllado, F., Martinez-Garcia, B., Vargas, M. & Diaz-Ruiz, J.R. Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 3, 3 (2003).
Aalto, A.P. et al. Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage phi6 RNA-dependent RNA polymerase. RNA 13, 422–429 (2007).
Huang, L. et al. Efficient and specific gene knockdown by small interfering RNAs produced in bacteria. Nat. Biotechnol. 31, 350–356 (2013).
Blau, J.A. & McManus, M.T. Renewable RNAi. Nat. Biotechnol. 31, 319–320 (2013).
Voinnet, O., Pinto, Y.M. & Baulcombe, D.C. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96, 14147–14152 (1999).
Silhavy, D. et al. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21, 3070–3080 (2002).
Ye, K., Malinina, L. & Patel, D.J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426, 874–878 (2003).
Jin, J., Cid, M., Poole, C.B. & McReynolds, L.A. Protein mediated miRNA detection and siRNA enrichment using p19. Biotechniques 48, xvii–xxiii (2010).
Xiao, J., Feehery, C.E., Tzertzinis, G. & Maina, C.V. E. coli RNase III(E38A) generates discrete-sized products from long dsRNA. RNA 15, 984–991 (2009).
Smith, D.B. & Corcoran, L.M. Expression and purification of glutathione-S-transferase fusion proteins. Curr. Protoc. Mol. Biol. 28, 16.7.1–16.7.7 (2001).
Henschel, A., Buchholz, F. & Habermann, B. DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res. 32, W113–W120 (2004).
Surendranath, V., Theis, M., Habermann, B.H. & Buchholz, F. Designing efficient and specific endoribonuclease-prepared siRNAs. Methods Mol. Biol. 942, 193–204 (2013).
Lee, S.K. et al. Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood 106, 818–826 (2005).
Boese, Q. et al. Mechanistic insights aid computational short interfering RNA design. Methods Enzymol. 392, 73–96 (2005).
Chen, P.Y. et al. Strand-specific 5′-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA 14, 263–274 (2008).
Jackson, A.L. et al. Position-specific chemical modification of siRNAs reduces 'off-target' transcript silencing. RNA 12, 1197–1205 (2006).
Jackson, A.L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003).
Robbins, M. et al. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol. Ther. 15, 1663–1669 (2007).
Acknowledgements
We thank J. Carrington (Donald Danforth Plant Science Center) for providing the p19 clone. We also thank J. Jin and L. McReynolds from New England Biolabs and P. Deighan and E. Kiner from Harvard Medical School for assistance and advice in establishing this method. We also thank Lieberman Laboratory members for technical assistance. This work was supported by a US National Institutes of Health grant (no. AI087431; J.L.) and a GlaxoSmithKline(GSK)-Immune Disease Institute(IDI) Alliance fellowship (L.H.).
Author information
Authors and Affiliations
Contributions
L.H. designed the protocol with suggestions from J.L. and L.H., and J.L. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Huang, L., Lieberman, J. Production of highly potent recombinant siRNAs in Escherichia coli. Nat Protoc 8, 2325–2336 (2013). https://doi.org/10.1038/nprot.2013.149
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.149
This article is cited by
-
Kaolinite nanoclay-shielded dsRNA drenching for management of Globodera pallida: An environmentally friendly pest management approach
Protoplasma (2024)
-
Negative feedback circuit for toll like receptor-8 activation in human embryonic Kidney 293 using outer membrane vesicle delivered bi-specific siRNA
BMC Immunology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.