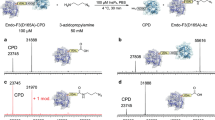
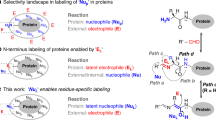
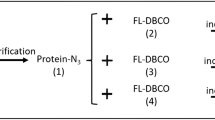
Abstract
This protocol describes the use of sortase-mediated reactions to label the N terminus of any given protein of interest. The sortase recognition sequence, LPXTG (for Staphylococcus aureus sortase A) or LPXTA (for Streptococcus pyogenes sortase A), can be appended to a variety of probes such as fluorophores, biotin or even to other proteins. The protein to be labeled acts as a nucleophile by attacking the intermediate formed between the probe containing the LPXTG/A motif and the sortase enzyme. If sortase, the protein of interest and a suitably functionalized label are available, the reactions usually require less than 3 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
06 December 2013
In the version of this article initially published, the genus names for the bacteria were transposed in the abstract so that they incorrectly read Streptococcus aureus and Staphylococcus pyogenes instead of Staphylococcus aureus and Streptococcus pyogenes. The error has been corrected in the HTML and PDF versions of the article.
References
Guimaraes, C.P. et al. Identification of host cell factors required for intoxication through use of modified cholera toxin. J. Cell Biol. 195, 751–764 (2011).
Gautier, A. et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 (2008).
Stephanopoulos, N. & Francis, M.B. Choosing an effective protein bioconjugation strategy. Nat. Chem. Biol. 7, 876–884 (2011).
Dirksen, A. & Dawson, P.E. Expanding the scope of chemoselective peptide ligations in chemical biology. Curr. Opin. Chem. Biol. 12, 760–766 (2008).
Dmochewitz, L. et al. A recombinant fusion toxin based on enzymatic inactive c3bot1 selectively targets macrophages. PLoS ONE 8, e54517 (2013).
Jang, J.I. et al. Expression and delivery of tetanus toxin fragment C fused to the N-terminal domain of SipB enhances specific immune responses in mice. Microbiol. Immunol. 56, 595–604 (2012).
Gellerman, G., Baskin, S., Galia, L., Gilad, Y. & Firer, M.A. Drug resistance to chlorambucil in murine B-cell leukemic cells is overcome by its conjugation to a targeting peptide. Anticancer Drugs 24, 112–119 (2013).
Baratova, L.A. et al. The topography of the surface of potato virus X: tritium planigraphy and immunological analysis. J. Gen. Virol. 73, 229–235 (1992).
Schwede, A., Jones, N., Engstler, M. & Carrington, M. The VSG C-terminal domain is inaccessible to antibodies on live trypanosomes. Mol. Biochem. Parasit. 175, 201–204 (2011).
Hess, G.T. et al. M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins. Bioconjugate Chem. 23, 1478–1487 (2012).
Reyes-Turcu, F.E. et al. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124, 1197–1208 (2006).
Love, K.R., Pandya, R.K., Spooner, E. & Ploegh, H.L. Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem. Biol. 4, 275–287 (2009).
Greenhalf, W., Stephan, C. & Chaudhuri, B. Role of mitochondria and C-terminal membrane anchor of Bcl-2 in Bax-induced growth arrest and mortality in Saccharomyces cerevisiae. FEBS Lett. 380, 169–175 (1996).
Kataoka, T. et al. Bcl-rambo, a novel Bcl-2 homologue that induces apoptosis via its unique C-terminal extension. J. Biol. Chem. 276, 19548–19554 (2001).
Miyadera, T. & Kosower, E.M. Receptor site labeling through functional groups. 2. Reactivity of maleimide groups. J. Med. Chem. 15, 534–537 (1972).
Palmer, M., Buchkremer, M., Valeva, A. & Bhakdi, S. Cysteine-specific radioiodination of proteins with fluorescein maleimide. Anal. Biochem. 253, 175–179 (1997).
Hnatowich, D.J. et al. Labeling peptides with technetium-99m using a bifunctional chelator of a N-hydroxysuccinimide ester of mercaptoacetyltriglycine. J. Nucl. Med. 39, 56–64 (1998).
Rabuka, D., Rush, J.S., deHart, G.W., Wu, P. & Bertozzi, C.R. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat. Protoc. 7, 1052–1067 (2012).
Wu, P. et al. Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proc. Natl. Acad. Sci. USA 106, 3000–3005 (2009).
Raschke, T.M., Kho, J. & Marqusee, S. Confirmation of the hierarchical folding of RNase H: a protein engineering study. Nat. Struct. Biol. 6, 825–831 (1999).
Rowe, S.M., Miller, S. & Sorscher, E.J. Cystic fibrosis. N. Engl. J. Med. 352, 1992–2001 (2005).
Carrico, Z.M. et al. N-Terminal labeling of filamentous phage to create cancer marker imaging agents. ACS Nano. 6, 6675–6680 (2012).
Guimaraes, C.P. et al. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc. 8, 1787–1799 (2013).
Paterson, G.K. & Mitchell, T.J. The biology of Gram-positive sortase enzymes. Trends Microbiol. 12, 89–95 (2004).
Ton-That, H., Liu, G., Mazmanian, S.K., Faull, K.F. & Schneewind, O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96, 12424–12429 (1999).
Mazmanian, S.K., Liu, G., Ton-That, H. & Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 (1999).
Spirig, T., Weiner, E.M. & Clubb, R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 82, 1044–1059 (2011).
Popp, M.W., Antos, J.M. & Ploegh, H.L. Site-specific protein labeling via sortase-mediated transpeptidation. Curr. Protoc. Protein Sci. 56, 15.3.1–15.3.9 (2009).
Popp, M.W. & Ploegh, H.L. Making and breaking peptide bonds: protein engineering using sortase. Angew Chem. Int. Ed. Engl. 50, 5024–5032 (2011).
Ton-That, H. & Schneewind, O. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J. Biol. Chem. 274, 24316–24320 (1999).
Race, P.R. et al. Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J. Biol. Chem. 284, 6924–6933 (2009).
Tanaka, T., Yamamoto, T., Tsukiji, S. & Nagamune, T. Site-specific protein modification on living cells catalyzed by sortase. Chembiochem 9, 802–807 (2008).
Yamamoto, T. & Nagamune, T. Expansion of the sortase-mediated labeling method for site-specific N-terminal labeling of cell surface proteins on living cells. Chem. Commun. 9, 1022–1024 (2009).
Tsukiji, S. & Nagamune, T. Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering. Chembiochem 10, 787–798 (2009).
Williamson, D.J., Fascione, M.A., Webb, M.E. & Turnbull, W.B. Efficient N-terminal labeling of proteins by use of sortase. Angew Chem. Int. Ed. Engl. 51, 9377–9380 (2012).
Cudic, P. & Stawikowski, M. Peptidomimetics: Fmoc solid-phase pseudopeptide synthesis. Methods Mol. Biol. 494, 223–246 (2008).
Witte, M.D. et al. Production of unnaturally linked chimeric proteins using a combination of sortase -catalyzed transpeptidation and click chemistry. Nat. Protoc. 8, 1808–1819 (2013).
Witte, M.D. et al. Preparation of unnatural N-to-N and C-to-C protein fusions. Proc. Natl. Acad. Sci. USA 109, 11993–11998 (2012).
Antos, J.M. et al. Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. J. Am. Chem. Soc. 131, 10800–10801 (2009).
Barrett, A.J., Rawlings, N.D. & Woessner, J.F. Handbook of Proteolytic Enzymes (Academic Press, 1998).
Liu, D., Xu, R., Dutta, K. & Cowburn, D. N-terminal cysteinyl proteins can be prepared using thrombin cleavage. FEBS Lett. 582, 1163–1167 (2008).
Gisin, B.F. The monitoring of reactions in solid-phase peptide synthesis with picric acid. Anal. Chim. Acta. 58, 248–249 (1972).
Vojkovsky, T. Detection of secondary amines on solid phase. Pept. Res. 8, 236–237 (1995).
Schagger, H. Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 (2006).
Hess, G.T., Guimaraes, C.P., Spooner, E., Ploegh, H.L. & Belcher, A.M. Orthogonal labeling of M13 minor capsid proteins with DNA to self-assemble end-to-end multiphage structures. ACS Synth Biol., doi:10.1021/sb400019s (2013).
Acknowledgements
This work was supported by funding from The Netherlands Organisation for Scientific Research (to M.D.W.) and the US National Institutes of Health (NIH; grant no. RO1 AI087879 to H.L.P.).
Author information
Authors and Affiliations
Contributions
C.S.T., C.P.G. and H.L.P. conceived of and drafted the manuscript. C.S.T., M.D.W., A.E.M.B., L.K. and C.P.G. participated in the optimization of the protocols and wrote the manuscript. H.L.P. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Theile, C., Witte, M., Blom, A. et al. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat Protoc 8, 1800–1807 (2013). https://doi.org/10.1038/nprot.2013.102
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.102
This article is cited by
-
Visualization of BOK pores independent of BAX and BAK reveals a similar mechanism with differing regulation
Cell Death & Differentiation (2023)
-
Site-specific bioorthogonal protein labelling by tetrazine ligation using endogenous β-amino acid dienophiles
Nature Chemistry (2023)
-
Structural and functional asymmetry of RING trimerization controls priming and extension events in TRIM5α autoubiquitylation
Nature Communications (2022)
-
Preclinical evaluation of [99mTc]Tc-labeled anti-EpCAM nanobody for EpCAM receptor expression imaging by immuno-SPECT/CT
European Journal of Nuclear Medicine and Molecular Imaging (2022)
-
Force spectroscopy of single cells using atomic force microscopy
Nature Reviews Methods Primers (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.