Abstract
Methods for site-specific modification of proteins should be quantitative and versatile with respect to the nature and size of the biological or chemical targets involved. They should require minimal modification of the target, and the underlying reactions should be completed in a reasonable amount of time under physiological conditions. Sortase-mediated transpeptidation reactions meet these criteria and are compatible with other labeling methods. Here we describe the expression and purification conditions for two sortase A enzymes that have different recognition sequences. We also provide a protocol that allows the functionalization of any given protein at its C terminus, or, for select proteins, at an internal site. The target protein is engineered with a sortase-recognition motif (LPXTG) at the place where modification is desired. Upon recognition, sortase cleaves the protein between the threonine and glycine residues, facilitating the attachment of an exogenously added oligoglycine peptide modified with the functional group of choice (e.g., fluorophore, biotin, protein or lipid). Expression and purification of sortase takes ∼3 d, and sortase-mediated reactions take only a few minutes, but reaction times can be extended to increase yields.
Similar content being viewed by others
Introduction
One of the goals of protein engineering is the installation of desirable features, template-encoded or otherwise, on proteins that naturally lack them. The ability to confer different functionalities onto a protein of interest enables a broad array of applications. Attachment of a fluorophore to a protein allows for its use in live-cell microscopy, whereas generation of a fusion between an antibody and a payload of interest, such as a toxin or an antigen, can find use in therapeutics and vaccine development, respectively1,2,3,4.
Several strategies based on genetic, chemical, enzymatic or chemo-enzymatic methods equip proteins with functional groups. Genetic engineering is the method of choice when modification at a precise site is required. However, the effect of the genetically appended sequence on expression, folding and function of the final product is difficult to predict. Some proteins are simply refractory to the construction of functional fusions by standard genetic means1. The range of modifications that can be applied to a protein as a fusion product is limited in the first instance to those that are template-encoded.
Chemical modifications of proteins are more versatile but lack precision, as they usually target exposed cysteine or lysine residues. Moreover, because the reactions often call for non-physiological reaction conditions (with respect to pH, reducing conditions and ionic composition), chemical damage of the target protein can occur. Proteins can be selectively labeled at their C terminus using the expressed protein ligation method, but generation of the required thioester from the protein-intein fusion is, in many cases, a low-yield process5. Only recently has this issue been addressed6.
Enzymatic methods can also overcome some of the drawbacks observed during chemical labeling, and they afford site-specific protein modification. However, these methods often require the following: genetic installation of sizable catalytically active protein domains (such as the O6-alkylguanine-DNA alkyltransferases (SNAP- or CLIP-based technology7,8) and haloalkane dehalogenases (HaloTag technology, 20–40 kDa; ref. 9)) onto the protein substrate; installation of the 15-aa BirA acceptor peptide10, the use of which is limited to biotin and its synthetically demanding chemical derivatives; engineering of a 13-aa acceptor peptide for lipoic acid ligase11, with the limitation that it primarily accepts lipid substrates and therefore mutant screens are required to incorporate new functionalities; the use of the formylglycine-generating enzyme that converts a cysteine residue within the context of a LCTPSR sequence (aldehyde tag) into formylglycine that can be used in oxime ligations12,13; or exploitation of the enzyme phosphopantotheinyltransferase to conjugate CoA-derived molecules to a specific 11-aa sequence14.
Sortase-mediated transpeptidation reactions are a versatile complement to these protein modification strategies15,16,17,18, and they predominantly rely on the use of modified peptides, readily accessible by solid-phase peptide synthesis using commercially available building blocks. Besides labeling of the protein of interest at its C terminus or at an internal region as described here, sortases allow functionalization of the N terminus19 and creation of non-natural fusions (i.e., N-N or C-C chimeras) via the installation of click handles20. By using sortases, we achieve labeling with similar precision to that afforded by genetic fusions, and we also provide ready access to protein derivatives and structures that are unattainable genetically.
Sortases and mechanism of action
Gram-positive bacteria display proteins at their surface to enable them to acquire nutrients, evade host immunity or adhere to sites of infection21. Sortases comprise a family of membrane-associated transpeptidases that anchor those proteins to the cell wall17,22. The different members of the sortase family can be divided into four subfamilies, on the basis of their distinct primary sequences and substrates23. Although sortases of type A accept a large number of protein substrates, sortases of types B–D have more specialized functions and therefore fewer substrates. In Staphylococcus aureus, proteins targeted to the bacterial surface have a conserved sortase-recognition motif, Leu-Pro-Xxx-Thr-Gly (LPXTG, where X is any amino acid and glycine cannot be a free carboxylate), at or near the C terminus. Upon recognition, sortase A cleaves between the threonine and glycine residues to form an acyl-enzyme intermediate. The active-site cysteine of sortase A forms a bond with the carbonyl of the threonine residue of the target protein. This intermediate is then resolved by nucleophilic attack by the free amino group of the cell wall precursor lipid II. This lipid II–linked protein conjugate is incorporated during cell wall synthesis, and consequently the protein is displayed at the surface24 (Fig. 1).
Adapted from ref. 24.
Diversity of applications
Sortase-mediated reactions are applicable to any protein of interest, provided it contains an LPXTG motif as the sortase target or a suitably exposed glycine residue to serve as the incoming nucleophile. Both modifications (LPXTG, glycine) can be introduced using standard molecular cloning protocols. In addition, sortases A are easily expressed in soluble recombinant form and in excellent yield in Escherichia coli. The natural nucleophile, lipid II, can be replaced by any peptide with an oligoglycine (Gly1−5) at the N terminus (in many cases a single glycine suffices). In turn, the peptides can be decorated with any molecule accessible through chemical synthesis (e.g., fluorophores, biotin, cross-linkers, lipids, carbohydrates, nucleic acids)1,16,17,25,26,27,28,29, provided that a free N-terminal glycine remains available on the peptide used as the incoming nucleophile. Thus, incubation of sortase, LPXTG-containing protein and nucleophile leads to the covalent attachment of that nucleophile to the protein of interest in a site-specific manner. Because the oligoglycine peptide that serves as the nucleophile is functionalized beforehand, the chemical reaction conditions used to incorporate the functional group damage neither the sortase nor the protein substrate as long as the modified oligoglycine peptide remains in solution once it is added to the sortase reaction. Proteins can also serve as nucleophiles, provided they display a suitably exposed (stretch of) glycine(s) at their N terminus (Gly1−5). Although in many cases one glycine is enough, we recommend installing more glycines, especially in those nucleophiles predicted to have a poorly exposed N terminus. The presence of even a single additional glycine can markedly change the efficiency of the reaction30. Such a modification allows the proteins to be N-terminally labeled with functionalized peptides30,31 or to form protein-protein adducts1,4,32.
By relying on a common mechanistic principle, sortagging affords ready access to a wealth of site-specific modifications: C-terminal16,25,33, internal loop regions1,34, N-terminal30,31 and formation of cyclized (poly)peptides30,35,36. We have mainly used sortases of type A derived from S. aureus and Streptococcus pyogenes. Versions of S. aureus with improved Kcat values37, as well as mutant versions that do not require Ca2+ ions38, have been reported, and they further extend the range of reaction conditions and applications. S. pyogenes sortase A accepts dialanine (poly)peptides as nucleophiles39. The possibility of using two orthogonal sortases increases the versatility of these labeling reactions, as one can attach two different labels to one and the same molecule of choice40.
Limitations
More than 50 different substrates including peptides28,35, soluble proteins16,30,33,34, membrane proteins displayed at the cell surface16,41, M13 bacteriophage42, budding influenza virus3, antibodies4,29,32, bacterial toxins1,40 and preassembled complexes1,40 have yielded to sortase labeling. Although we have yet to encounter a protein that could not be labeled using sortase, a parameter that critically influences the efficiency of the labeling reaction is the flexibility and accessibility of the region to be labeled. In most cases, this issue can be circumvented, yet to some extent the intrinsic structure of the substrate protein will impose limitations. This is true in particular when labeling an internal region of the protein, as some loop regions may not tolerate any type of genetic engineering.
One initial limitation of using sortase A from S. aureus was its obligate Ca2+-dependent activity16. The presence of calcium in the reaction buffer precludes the use of phosphate-based buffers. Sortase A from S. pyogenes39 or from a mutant form of S. aureus (E105K/E108A (ref. 38)) is Ca2+ independent and thus circumvents this limitation.
Not every laboratory is equipped to perform peptide synthesis, and commercial vendors provide such services. To assist those interested in synthesizing their own peptides, we have included protocols that describe the synthesis of probes of general utility (biotin and fluorophores). It requires minimal specialized equipment (reaction vessels for peptide synthesis) and involves reactions readily executed in a laboratory outfitted for biochemical work (fume hoods, appropriate organic waste disposal and a lyophilizer).
Advantages of the method
Sortase-mediated reactions are site-specific; afford high labeling yields; are versatile with respect to the moieties to be attached to the protein of interest (biotin1,16, fluorophores1,3,40, reporter peptides1, sugars27, lipids25 and so on); are flexible regarding the labeling position on a protein: N, C, both N and C terminus, and solvent-accessible loops; enable the cyclization of proteins or peptides, if N and C termini are in close proximity; can be carried out under physiological conditions; require minimal modification of the target protein (5-amino acid recognition sequence); can be orthogonal, as sortases recognizing different motives are available; and can be performed using sortase A in solution or sortase A immobilized on a solid support43.
Experimental design
The components of any sortagging reaction are: sortase, substrate and nucleophile.
Expression and production of sortase A. We have three different sortases available: the Ca2+-dependent sortase A from S. aureus (the first one to be described and the most commonly used), its mutant version (E105K/E108A (ref. 38)) and sortase A from S. pyogenes; the latter two are Ca2+ independent. Because sortase is a membrane protein in Gram-positive bacteria, we use versions in which the transmembrane domain has been eliminated and replaced with a hexahistidine (His6) purification tag. Two soluble versions exist for S. aureus wild-type sortase A, with an N-terminal deletion of either 25 aa (ref. 24) or 59 aa. The enzymatic activity of both versions is identical44, but their molecular weight is different. This is a useful trait to explore in those cases where the molecular weights of the protein to be labeled and of the sortase to be used are similar. In addition, we equipped the Δ59 truncated version with a thrombin cleavage site that releases the His6 tag upon digestion. This not only facilitates further downstream purification but also increases the mobility of sortase in SDS-PAGE gels, allowing a clear-cut distinction between sortase and substrate if required. The same protocol can be used to express and purify the various sortases. Sortase A is expressed in E. coli as a His6 fusion to facilitate purification using a nickel-chelate affinity column. The described standard procedure yields ∼40 mg l−1 S. aureus sortase A, 25 mg l−1 Ca2+-independent S. aureus sortase A and 30 mg l−1 S. pyogenes sortase A in good purity. The protein also remains soluble when it is concentrated.
Engineering substrates for C-terminal labeling. Substrates equipped with the LPXTG-recognition motif for S. aureus, or LPXTA for S. pyogenes, can be engineered using standard molecular cloning protocols. Although any amino acid can precede the threonine residue, a glutamic acid is often used because it is commonly found in the natural sortase A substrates45. The sortase-recognition sequence is usually engineered at the C terminus of the protein to be modified, with the G or A residue in amide linkage, followed by an affinity purification handle (e.g., His6) that is lost upon reaction (Fig. 2). The efficiency of the sortase-mediated reaction depends on the flexibility and accessibility of the region comprising the sortase A-recognition motif. Thus, if the C terminus of the protein is known to be hidden (in the absence of a known structure, a failure to yield to sortase labeling would be a clear indication of lack of accessibility), we recommend engineering a flexible linker composed of (Gly4Ser)n preceding the LPXTG/A sequence. The length of such linkers needs to be tested empirically for each protein of interest. If the amino acid immediately following the initial methionine is a glycine or alanine residue, you should consider deleting the amino acid or mutating it (to serine, for example). Having an LPXTG/A-recognition sequence at the C terminus and glycine/alanine at the N terminus of the same protein substrate may result in cyclization or polymerization of the protein as a competing side reaction during labeling30.
A protein modified at its C terminus with the LPXTG sortase-recognition motif followed by a handle (usually His6) is incubated with S. aureus Sortase A. Sortase cleaves the threonine-glycine bond and via its active site cysteine residue forms an acyl intermediate with threonine in the protein. Addition of a peptide probe comprising a series of N-terminal glycine residues and a functional moiety of choice resolves the intermediate, thus regenerating the active site cysteine (HS) on sortase and ligating the peptide probe to the C terminus of the protein.
Engineering substrates for internal loop labeling. Site-specific modification of an internal solvent-exposed region in the protein of interest is a particular case of C-terminal labeling. As long as the LPXTG/A motif is introduced in an unstructured segment of the substrate protein, sortase can recognize the sequence34. An LPXTG motif that is highly structured is usually a poor sortase substrate1,34. Flexibility can be ensured through installation of a specific protease cleavage site, immediately downstream of the sortase motif1. Upon cleavage, the newly exposed C terminus including the LPXTG/A motif is likely to be unstructured. Depending on the sequence of the protein, we rely on established site-directed mutagenesis strategies to insert an LPXTG/A motif at the intended site. As sortase cleaves the protein at the site of recognition, it is likely that the two halves of the protein will separate upon sortagging unless otherwise stabilized, for example, through a disulfide bond1 or for topological reasons34. Thus, selecting an internal location for a sortase site in a loop region constrained by a disulfide bond might minimize the risk of such disintegration (Fig. 3). Note that the protease to use must be tested empirically to ensure that the protease does not cleave elsewhere within the protein. Trypsin and Factor Xa are examples of proteases used for this purpose1.
A protein comprising a loop formed by the establishment of a disulfide bond is modified to contain the sortase-recognition motif (LPXTG), followed by a specific protease cleavage site. To increase flexibility of the LPXTG-containing region, the loop is nicked with the protease of choice, and the sortase-mediated reaction follows as described for C-terminal labeling. Note that as long as the region containing the LPXTG motif is flexible and accessible, a proteolytic event may not be required. The presence of a disulfide bridge is also not crucial for the reaction. Although such a bond is convenient to ensure that the protein will not disintegrate upon labeling, at times the topology and conformation of the protein of interest naturally ensures integrity.
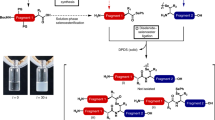
Peptide synthesis. Here we describe the manual synthesis of several peptides that can be used in reactions mediated by sortase A from S. aureus, and therefore contain glycine residues at the N terminus. For reactions using sortase A from S. pyogenes, an alanine-based peptide is used as a nucleophile. Their synthesis follows the same protocol, except for the use of Fmoc-Ala-OH in place of Fmoc-Gly-OH. Perform two or three repeated couplings of Fmoc-Ala-OH to obtain the di- and tri-alanine sequences, respectively. In addition, Fmoc-Lys(biotin)-OH, Fmoc-Lys(5-TAMRA)-OH and other conjugates can be purchased as premade building blocks. These building blocks are more expensive, but they reduce the time required for synthesis and facilitate purification of the desired final product.
Purification of the substrate-labeled product. Because the protein substrate is constructed with a His6 handle downstream of the LPXTG/A motif, the protein substrate molecules that were not labeled will retain the His6 tag upon reaction. The sortases are themselves tagged with His6. Thus, a convenient strategy to separate the labeled product from sortase and unreacted substrate is the use of affinity chromatography (Nickel–nitrilotriacetic acid (Ni-NTA) beads), followed by fast protein liquid chromatography (FLPC) or by a desalting column to remove the unreacted peptide nucleophile. Be sure to check compatibility of the incorporated functionality with Ni-NTA purification. We have noted that some functional groups, including acylhydrazones, bind to the resin.
Materials
REAGENTS
Caution
For all items marked with a 'Caution' callout, please use proper personal protection equipment (gloves, eye protection and proper attire). The use of these chemicals should be carried out in a fume hood when possible. For more information, please refer to each item's MSDS.
-
pQE30 sortase A S. aureus (Δ25), available upon request from H.L.P.24
-
pET28a sortase A S. aureus (Δ59), available upon request from H.L.P.
-
pET28a sortase A S. aureus (Δ59, E105K, E108A, Ca2+ independent), available upon request from H.L.P.
-
pET28a sortase A S. pyogenes (Δ81), available upon request from H.L.P.
-
E. coli BL21(DE3) (Invitrogen, C6000-03)
-
Kanamycin (Fisher Scientific, cat. no. BP906-5)
-
Ampicillin (Sigma-Aldrich, cat. no. A0166)
-
Luria-Bertani (LB) medium (Gentaur, cat. no. SD7002(S518)) and LB plates (BD, cat. no. 244510) (ampicillin 100 μg ml−1 or kanamycin 30 μg ml−1 for pQE30 or pET28-derived constructs, respectively)
-
Isopropyl-β-D-thiogalactopyranoside (Chem-Impex International, cat. no. 00194)
-
Nickel-nitrilotriacetic acid (Ni-NTA)-agarose resin (Qiagen, cat. no. 1018240)
-
Lysozyme (Sigma-Aldrich, cat. no. L6876)
-
DNase I (Roche, cat. no. 10 104 159 001)
-
Imidazole (Alfa Aesar, cat. no. A10221)
-
Common reagents for SDS-PAGE analysis
-
Loading LDS-buffer, 4× (Invitrogen, cat. no. NP0008)
-
Brilliant blue R (Sigma-Aldrich, cat. no. B7920)
-
Methanol (EMD, cat. no. MX0488-1)
Methanol is flammable and toxic.
-
Acetic acid (VWR, cat. no. BDH3094)
Acetic acid is flammable and corrosive.
-
Hydrochloric acid (HCl; EMD, cat. no. HX0603-4))
HCl is flammable, corrosive and toxic. Avoid skin contact and inhalation.
-
Ethanol (Pharmco-AAPER, cat. no. 111000190)
Ethanol is flammable.
-
Bicinchoninic acid (BCA)-protein reagent assay kit (Pierce, cat. no. 23227)
-
N,N-dimethylformamide (DMF; Applied Biosystems, cat. no. GEN002007)
DMF is flammable and toxic.
-
Acetonitrile (JT Baker Analytical, cat. no. 9017-03)
Acetonitrile is flammable and toxic.
-
N-methyl-2-pyrrolidone (NMP; Sigma-Aldrich, cat. no. 328634-2L)
NMP is flammable, an irritant and toxic.
-
Diisopropylethylamine (DIPEA; Fisher BioReagent, cat. no. BP592500)
DIPEA is highly flammable and corrosive.
-
Dichloromethane (DCM; VWR, cat. no. JT9305-3)
DCM is a carcinogen.
-
DMSO (EMD Chemicals, cat. no. MX1458-6)
DMSO is an irritant and flammable.
-
Diethyl ether (EMD Chemicals, cat. no. EX0185-8)
Diethyl ether is highly flammable.
-
Trifluoroacetic acid (TFA; Sigma-Aldrich, cat. no. T6508)
TFA is strongly corrosive and toxic.
-
Triisopropylsilane (TIS; Sigma-Aldrich, cat. no. 233781)
TIS is flammable.
-
Ethanedithiol (Sigma-Aldrich, cat. no. w348406)
Ethanedithiol is flammable and toxic.
-
5(6)-Carboxy-tetramethylrhodamine (TAMRA; Novabiochem, cat. no. 815030)
-
Biotin (Sigma-Aldrich, cat. no. B4501)
-
Fmoc-Lys(Mtt)-OH (EMD Biosciences, cat. no. 04-12-1137)
-
Fmoc-Cys(Trt)-OH (Novabiochem, cat. no. 852008)
-
Fmoc-Gly3-OH (Chem-Impex International, cat. no. 08072)
-
Fmoc-Gly-OH (Novabiochem, cat. no. 852001)
-
Fmoc-Ala-OH (Novabiochem, cat. no. 852003)
-
Fmoc-Lys(biotin)-OH (Novabiochem, cat. no. 852097)
-
Fmoc-Lys(5-TAMRA)-OH (AAT Bioquest, cat. no. 5045)
-
2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU; Novabiochem, cat. no. 851006)
HBTU is an irritant and is harmful.
-
Benzotriazol-1-yl-oxytripyrrolidinophosphoniumhexafluorophosphate (PyBOP; Novabiochem, cat. no. 851009)
This compound is an irritant and is harmful.
-
Rink amide resin SS, 100–200 mesh, 1% (wt/wt) divinylbenzene (DVB) (Advanced Chemtech, cat. no. SA5030)
-
tert-Butanol (Aldrich, cat. no. 36,053-8)
tert-Butanol is flammable.
-
Piperidine
-
NaCl
-
MgCl2
-
Glycerol
-
CaCl2
-
PBS
-
Maleimide
EQUIPMENT
-
Water bath
-
Bacterial shaker
-
Floor centrifuge capable of spinning 1-liter bacterial cultures at 6,000g and spinning 50-ml tubes at 20,000g
-
Motorized mechanical pipette
-
French Press for lysis of bacterial cells
-
Poly-Prep chromatography column (Bio-Rad)
-
FPLC (Äkta system or similar; GE Healthcare)
-
HiLoad 16/60 Superdex 75 prep grade size exclusion chromatography column
-
Amicon Ultra concentrators, 10 kDa NMWL (Millipore)
-
Polypropylene conical tubes, 50 ml (Corning or similar)
-
Micropipettes (5–1,000 μl) and respective tips
-
Centrifuge tubes, 1.5 ml
-
Test tube racks
-
End-over-end shaker
-
Heating block
-
Desalting PD-10 columns (GE Healthcare, cat. no. 17-0851-01)
-
Syringe and filter frit, 3 ml (New England Peptide, cat. no. AC0-003)
-
Screw cap glass peptide column with a frit filter bottom
-
Wrist-Action shaker (St. John Associates)
-
Swing bucket centrifuge (Beckman)
-
HPLC system (Agilent 1100 series)
-
Reverse-phase 1 × 25 cm protein and peptide C18 column (Vydac, cat. no. 218TP1010)
-
Instruments and consumables for liquid chromatography/mass spectrometry (LC/MS)
-
NMR spectrometer
-
Vacuum line
-
Lyophilizer
-
Glass pipettes and Pasteur pipettes
-
Syringes
-
Graduated cylinders
-
Pipette bulbs
-
Rotary evaporator (Büchi)
REAGENT SETUP
Piperidine in NMP, 20% (vol/vol)
-
Mix 20 ml of piperidine with 80 ml of NMP. The Fmoc deprotection solution can be stored at room temperature (RT; 21–25 °C) for 1 month.
Cleavage cocktail (95% (vol/vol) TFA, 2.5% (vol/vol) H2O and 2.5% (vol/vol) TIS)
-
Mix 4.75 ml of TFA, 125 μl of H2O and 125 μl of TIS. The cleavage cocktail should be freshly prepared for each experiment.
LB medium
-
Autoclave the medium. Store it at 4 °C for up to 6 months. Add the desired antibiotic just before using the medium.
LB medium agar plates
-
Prepare LB medium according to instructions. Autoclave the medium, allow it to cool down, and then add the desired antibiotic. Pour the medium into plates and store it at 4 °C for up to 3 months.
Kanamycin
-
Prepare kanamycin at a concentration of 30 mg ml−1 in water and filter-sterilize it.
Critical
Store it at −20 °C (1,000× stock) for up to 6 months.
Ampicillin, sodium salt
-
Prepare ampicillin at a concentration of 100 mg ml−1 in water and filter-sterilize it.
Critical
Store it at −20 °C (1,000× stock) for up to 6 months.
Isopropyl-β-D-thiogalactopyranoside
-
Prepare isopropyl-β-D-thiogalactopyranoside at a concentration of 0.5 M in water.
Critical
Store it at −20 °C (1,000× stock) for up to 1 month.
Imidazole, 1 M
-
Prepare imidazole in nickel-binding buffer and adjust the pH to 7.8 with HCl. Dilute the imidazole stock solution in nickel-binding buffer to make the lysis, wash and elution buffers as indicated. Store imidazole at 4 °C for up to 3 months.
Lysozyme
-
Store the powder at −20 °C for up to 1 year and dissolve it at a concentration of 10 mg ml−1 in water just before use (1,000× stock).
DNase
-
Store the powder at 4 °C for up to 1 year and dissolve it at 10 mg ml−1 in water just before use (1,000× stock).
Nickel-binding buffer
-
Nickel-binding buffer contains 50 mM Tris-HCl, pH 7.5, and 150 mM NaCl. Filter-sterilize the buffer and store it at 4 °C for up to 3 months.
Lysis buffer
-
Lysis buffer contains 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10 mM imidazole, 10% (vol/vol) glycerol, 1 mg ml−1 DNase and 1 mg ml−1 lysozyme. Filter-sterilize the buffer and store it at 4 °C for up to 3 months.
Wash buffer
-
Wash buffer contains 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 10 mM imidazole and 10% (vol/vol) glycerol. Filter-sterilize the buffer and store it at 4 °C for up to 3 months.
Elution buffer
-
Elution buffer contains 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 500 mM imidazole and 10% (vol/vol) glycerol. Filter-sterilize the buffer and store it at 4 °C for up to 3 months.
Coomassie blue staining
-
Dissolve 1.25 g of Brilliant Blue R in a mixture of methanol (200 ml), water (250 ml) and acetic acid (50 ml). Store it in a dark container at RT for up to 1 year.
Destaining solution
-
Mix water, ethanol and acetic acid in a ratio of 6:3:1. Store the solution at RT for up to 1 year.
Sortase buffer
-
Sortase buffer contains 500 mM Tris-HCl (pH 7.5), 1.5 M NaCl and 100 mM CaCl2 (not required if you are using a Ca2+-independent sortase; 10× stock). Filter-sterilize the buffer and store it at 4 °C for up to 3 months.
Purified sortase A in buffer
-
Use as described in Step 16.
Critical
Do not use a phosphate-based buffer if a Ca2+-dependent sortase A is used.
Purified target protein in buffer
-
Designed as indicated in the Experimental design section, this protein should be expressed and purified on the basis of the intrinsic nature of the protein or by following the same guidelines described for expression and purification of sortase.
Critical
Do not use a phosphate-based buffer if a Ca2+-dependent sortase A is used.
Oligoglycine peptide
-
Prepare oligoglycine peptide (if using S. aureus sortase A) or dialanine or trialanine peptide (if using S. pyogenes sortase A) stock solution: 10 mM in DMSO or water (10× stock). If an oligoglycine protein is used as the nucleophile, then dissolve it in buffer.
Critical
Do not use a phosphate-based buffer if a Ca2+-dependent sortase A is used.
EQUIPMENT SETUP
HPLC
-
LC−electrospray ionization (ESI)-MS analysis is done with a Micromass LCT mass spectrometer (Micromass MS Technologies) and a Paradigm MG4 HPLC system equipped with a HTC PAL autosampler (Michrom BioResources) and a Waters Symmetry 5-μm C8 column (2.1 × 50 mm, MeCN:H2O (0.1% (vol/vol) formic acid) gradient mobile phase, 150 μl min−1).
LC/MS
-
Peptides are purified using an Agilent 1100 Series HPLC system equipped with a Waters Delta Pak 15-μm, 100-Å C18 column (3 ml min−1). Detection is at 214 nm and 254 nm.
Procedure
Peptide synthesis
-
1
There are three options for synthesizing the peptides. Note that in all three options NMP may be replaced with DMF. Option A describes the procedure for the preparation of GGGK-biotin and GGGK-TAMRA peptides. In-solution coupling of N-hydroxysuccinimide (NHS) esters to (partially) protected peptides provides a route to couple base/acid-labile and/or more costly dyes (such as Alexa Fluor 647) or other suitable precursors to a GGG-containing peptide (GGGK-NHS ester, option B). An alternative for the installation of chemical groups of interest onto a peptide is to use a cysteine-maleimide reaction (option C). The procedure for option C is similar to that for the GGGK–NHS ester peptide, except that the lysine is replaced by a cysteine and a maleimide-derived dye or probe is substituted for the NHS ester. Under appropriate conditions, the maleimide will react with the cysteine exclusively, and therefore fully deprotected peptides can be used in option C.
-
A
GGGK-biotin and GGGK-TAMRA peptides
-
i
Resin preparation (15 min). Add 100 μmol of Rink amide resin (167 mg, 0.6 mmol g−1) into a capped glass column with a fritted glass bottom, solvate the resin in DCM (7 ml) by shaking it for 15 min in a wrist-action shaker at RT and remove the DCM by vacuum filtration.
-
ii
Deprotection and washing (30 min). Add 20% (vol/vol) piperidine solution in NMP (7 ml) and shake it for 15 min at RT to remove the resin's Fmoc-protecting groups.
-
iii
Remove the piperidine solution by vacuum filtration and wash the resin three times with NMP (7 ml, 1 min), three times with DCM (7 ml, 1 min) and an additional time with NMP (7 ml, 1 min).
-
iv
Coupling reactions and washing (2–3 h per coupling until Pause Point, 3.5 h total). Dissolve Fmoc-Lys(Mtt)-OH (188 mg, 300 μmol), HBTU (114 mg, 300 μmol) and DIPEA (104 μl, 600 μmol) in NMP (7 ml) and add it to the resin. Shake the suspension for 2 h at RT.
-
v
Remove the reaction solution by vacuum filtration and wash the resin three times with NMP (7 ml, 1 min) and three times with DCM (7 ml, 1 min). Confirm the coupling reaction by performing a Kaiser test or a microcleavage test (Box 1).
Critical Step
If the reaction is incomplete, repeat Step 1A(iv,v) with half the amount of reagents used for a standard coupling and shake it for 1 h at RT.
Critical Step
Only store protected peptides.
Pause point
Dry the resin under vacuum and store it at 4 °C.
-
vi
Selectively remove the 4-methyl trityl (Mtt)–protecting group with a solution of 97% (vol/vol) DCM, 2% (vol/vol) TIS and 1% (vol/vol) TFA. Shake the peptide resin with the cleavage solution (5 ml) for 30 min. Remove the solution by vacuum filtration, add an additional 5 ml of cleavage solution and shake it for 30 min to ensure complete removal of the Mtt. The side chain deprotected peptide remains attached to the resin.
Critical Step
The cleaved Mtt produces an orange color. Eventually, the color will fade from reaction with the TIS. If the color remains after the second 30-min reaction, perform a third 30-min deprotection.
-
vii
Wash the resin three times with NMP (7 ml, 1 min), three times with DCM (7 ml, 1 min), and then once briefly with NMP (7 ml, 1 min) containing DIPEA (500 μmol, 5 equiv.).
Critical Step
This final wash step neutralizes the resin and improves the coupling efficiency in the next step. This wash step should be brief. Prolonged exposure to alkaline conditions may result in partial removal of the Fmoc-protecting group.
-
viii
To synthesize the biotin-containing probe, make a solution of biotin (74 mg, 300 μmol), HBTU (114 mg, 300 μmol) and DIPEA (104 μl, 600 μmol) in NMP (7 ml). Add the solution to the resin and shake it for 2 h at RT. The reaction product consists of the G3K-biotin peptide. To synthesize the TAMRA-containing probe, prepare a solution of 5(6)-TAMRA (52 mg, 120 μmol), PyBOP (63 mg, 120 μmol) and DIPEA (42 μl, 240 μmol) in NMP (7 ml). Add the solution to the resin and shake it overnight at RT. To prevent photobleaching, avoid exposure to light where possible and wrap the column in aluminum foil.
-
ix
Repeat Step 1A(v) to monitor biotin/TAMRA coupling.
-
x
Repeat Step 1A(ii–v), except use Fmoc-Gly3-OH (123 mg, 300 μmol) instead of the lysine residue.
Critical Step
Three repeated couplings of Fmoc-Gly-OH can be used instead of Fmoc-Gly3-OH.
-
xi
Remove the terminal Fmoc group as indicated in Step 1A(ii,iii).
-
xii
Cleavage of the peptide from resin (3 h). Wash the residue an additional two times with DCM. Then, suspend the resin in a solution consisting of 95% (vol/vol) TFA, 2.5% (vol/vol) H2O and 2.5% (vol/vol) TIS (5 ml) for 2 h at RT.
-
xiii
Elute the cleavage solution into 90 ml of ice-cold (−20 °C) diethyl ether and rinse the resin with an additional 3 ml of the cleavage solution into the ether.
-
xiv
Store the ether solution at −20 °C for 20 min to precipitate the peptide. Centrifuge the suspension at 1,900g for 15 min at 4 °C. Decant the supernatant and gently evaporate the remaining ether under reduced pressure.
Caution
Because of the very flammable and volatile nature of diethyl ether, use a spark-free freezer and centrifuge.
Pause point
The crude product needs to be purified as soon as possible, but it can be stored as a solid at −20 °C for 1 week before purification. Lyophilize the peptide if you are going to store the crude peptide for a longer period.
-
xv
Verify the identity and purity by LC/MS analysis (linear gradient 5–45% (vol/vol) acetonitrile in 10 min) and NMR spectroscopy. If LC/MS shows that the crude peptide is of sufficient purity, HPLC purification may be omitted and the peptide may be used directly in sortase reactions.
-
xvi
HPLC purification (timing depends on amount of peptide and column used). Dissolve the dried peptide in water (2 ml) and centrifuge it at 16,000g for 10 min at RT. Up to 50% of tert-Butanol may be added to peptides that do not readily dissolve in pure water.
-
xvii
Purify the centrifuged supernatant by reverse-phase HPLC on a C18 column using a 10–70% water-acetonitrile gradient over 15 min with 0.1% (vol/vol) TFA, followed by flushing at 90% (vol/vol) acetonitrile for 5 min. We recommend starting with a small 100-μl injection and adjusting the gradient accordingly for peptide purity and ease of separation. Once a good gradient is established, we recommend purifying the rest of the crude material with 400–600-μl injections.
-
xviii
Analyze the fractions for the presence of the desired product by LC/MS (Step 1A(xv)), and lyophilize the desired fractions to dryness.
Critical Step
TAMRA-containing probes consist of a mixture of regioisomers, which are easily separated by reverse-phase HPLC. Both regioisomers can be used in sortase A–catalyzed transacylation reactions.
Pause point
The lyophilized peptide can be stored at −20 °C indefinitely.
Timing 2–3 d
-
i
-
B
GGGK–NHS ester probes
-
i
Follow Step 1A(ii–v) but by using Fmoc-Lys(Boc)-OH (141 mg, 300 μmol) instead of the Mtt-protected lysine.
-
ii
Follow Step 1A(ii–v) by substituting Fmoc-Gly3-OH (123 mg, 300 μmol) for the lysine residue.
-
iii
Cleave and precipitate the peptide from the resin with the Fmoc group still on the terminal glycine, as described in Step 1A(xii–xv).
Critical Step
It is crucial to leave the Fmoc-protecting group on the peptide, as it prevents unwanted side reactions during the NHS ester coupling.
Critical Step
Hydrophobic peptides may precipitate poorly. Dry the supernatant of the ether precipitation by rotary evaporation to increase the yield.
Pause point
Store the crude peptide at −20 °C for up to 1 month. If it is lyophilized, store it at −20 °C indefinitely.
-
iv
Purify the peptide by reverse-phase HPLC as indicated in Step 1A(xvi,xvii). If LC/MS shows that the crude peptide is of sufficient purity, this HPLC purification may be omitted. tert-Butanol may be added to peptides that do not dissolve in pure H2O before HPLC purification.
-
v
NHS ester coupling. Dissolve the peptide in DMSO.
-
vi
Add three to five equivalents of the Fmoc-GGGK peptide in DMSO to the NHS ester and add five equivalents of DIPEA.
-
vii
Incubate the reaction for 16 h at RT. To prevent photobleaching or photoconversions, exclude the peptide from light by covering its containers in aluminum foil.
Critical Step
It is important to leave the reaction for 16 h. Although the coupling of the NHS ester may be faster, the added DIPEA also facilitates (slow) removal of the Fmoc-protecting group.
-
viii
Acidify the reaction mixture with 0.1% (vol/vol) aqueous TFA and purify the peptide by reverse-phase HPLC, as described in Step 1A(xvi–xviii).
Pause point
The crude product should be purified as soon as possible, but it can be stored as a solid at −20 °C for a week before purification. The pure peptide can be stored at −20 °C indefinitely.
Timing 3 d
-
i
-
C
GGGC–maleimide-containing probes
-
i
Perform Step 1A(i–v) with Fmoc-Cys(Trt)-OH to load the resin.
-
ii
Perform Step 1A(ii–v) using Fmoc-Gly-Gly-Gly-OH.
-
iii
Perform Step 1A(ii,iii) to remove the N-terminal Fmoc-protecting group.
-
iv
Cleave the peptide from the resin by incubating the resin twice with 1 ml of a cleavage cocktail containing 93% (vol/vol) TFA, 2.5% (vol/vol) TIS, 2% (vol/vol) ethanedithiol and 2.5% (vol/vol) water for 2 h.
Critical Step
Addition of ethanedithiol prevents the formation of disulfide bridges.
-
v
Precipitate and collect the peptide as described in Step 1A(xiii,xiv).
-
vi
Remove the supernatant and dissolve the pellet in 2 ml of methanol.
-
vii
Precipitate and collect the peptide as described in Step 1A(xiii,xiv).
Critical Step
Removal of residual of ethanedithiol is important, as even trace amounts can interfere in the maleimide-thiol reaction.
-
viii
Analyze, purify (if needed) and lyophilize the crude H2N-GGGC-CONH2 as described in Step 1A(xvii).
Pause point
Store the lyophilized peptide at −20 °C until it is needed.
-
ix
Maleimide-thiol coupling. Dissolve the peptide (2 equiv., typically 80 μmol) in PBS (0.25 ml) and add maleimide (1 equiv.) in DMF or DMSO (0.25 ml). Note that two equivalents of peptide are added to completely consume precious maleimide-dye conjugates. Smaller amounts of peptide can be used for less expensive/precious maleimides. The use of less peptide will simplify purification.
-
x
Incubate the mixture overnight at RT and subsequently quench the reaction with 0.1% (vol/vol) aqueous TFA (2 ml).
-
xi
Purify, analyze and lyophilize the product as described in Step 1A(xv–xviii).
Critical Step
Prolonged storage of the probe in aqueous solution can result in hydrolysis of the maleimide. The resulting ring-opened product is still an excellent probe for the sortase reaction, but it may cause difficulty during ion-exchange purification of the sortagged proteins. It is therefore crucial that probes containing maleimide-coupled functional groups be stored as lyophilized powders.
Pause point
The lyophilized product can be stored at −20 °C as powder indefinitely.
-
xii
Verify the identity and purity by LC/MS analysis (linear gradient 5–45% (vol/vol) acetonitrile in 10 min) and NMR spectroscopy.
Timing 3 d
-
i
-
A
Expression and production of sortase A
Timing 3 d
-
2
Transform competent BL21(DE3) E. coli with pET28a S. aureus sortase A (Δ59), pET28a Ca2+-independent S. aureus sortase A (Δ59) or pET28a S. pyogenes sortase A onto LB-kanamycin plates or E. coli with pQE30 S. aureus sortase A (Δ25) onto LB-ampicillin plates, and then incubate the plates at 37 °C overnight.
-
3
Start overnight culture. Inoculate an individual colony from the plate into 100 ml of LB-ampicillin or LB-kanamycin medium (depending on the construct) and shake it at 220 r.p.m. at 37 °C overnight.
Critical Step
Use freshly transformed plates to ensure optimal expression.
-
4
Start expression culture. Dilute 10 ml of the overnight culture into 1 liter of LB medium with the corresponding antibiotic. Shake it at 37 °C until the measured absorbance at 600 nm (A600) is 0.4–0.6 (∼3 h). Reserve a 100-μl sample of the culture as the preinduction control for testing sortase expression.
-
5
Induce protein expression. Add Isopropyl-β-D-thiogalactopyranoside to a final concentration of 0.5 mM and shake it at 25 °C for 16 h.
-
6
Collection of bacteria. Reserve a 100-μl sample of the culture as the postinduction control for testing sortase expression. Centrifuge the culture at 6,000g for 15 min at 4 °C. Decant the supernatant. Resuspend the pellet in 50 ml of nickel-binding buffer, transfer it to a 50-ml centrifuge tube using a motorized pipette and centrifuge the pellet again.
Pause point
The pellet can be frozen at −80 °C and processed when convenient. We advise analyzing the pre- and post-induction samples by SDS-PAGE before proceeding to the purification step.
Purification and storage of sortase A
Timing 1–2 d
-
7
Lyse the bacteria in 25 ml of ice-cold lysis buffer and transfer the mixture to the chamber of a prechilled French press. Lyse the cells under a pressure of 1,000 p.s.i. Allow the bacterial lysate to cool on ice for 2 min and repeat the process. Clarify the cell lysate at 20,000g for 30 min at 4 °C. Keep the supernatant on ice.
-
8
Pack a Poly-Prep column with 1–1.5 ml (bed volume) of Ni-NTA agarose resin and wash it with 10 column-volumes of wash buffer (by gravity flow).
Critical Step
Do not allow the column to run dry.
-
9
Load the bacterial supernatant onto the column by gravity flow.
-
10
Wash the column with 50 column-volumes of wash buffer to remove nonspecifically bound proteins.
-
11
Elute with three column-volumes of ice-cold elution (high-imidazole) buffer.
-
12
Analyze a sample of the eluate by SDS-PAGE.
-
13
The sortase preparation is best further purified by gel filtration. This affords removal of the remaining contaminants and of the imidazole contained in the elution buffer. Pre-equilibrate a Hi-Load Superdex 75 16/60 column with 40 ml of nickel-binding buffer (no imidazole), at a rate of 1 ml min−1.
-
14
Inject the eluted protein sample (4–5 mg) and maintain a flow rate of 1 ml min−1. Collect 1.3-ml fractions.
-
15
On the basis of the UV spectra, identify and collect the protein-containing fractions and analyze them by SDS-PAGE. Pool the fractions that contain pure sortase and concentrate them to 25 mg ml−1 using an Amicon Ultra concentrator. The protein concentration is determined by BCA assay. An aliquot of the protein is analyzed by SDS-PAGE to check for purity.
Critical Step
We often detect the presence of contaminants in the preparation of the S. pyogenes sortase A. These contaminants do not affect the activity of the enzyme, but the enzyme preparation can be further purified by anion-exchange chromatography (Mono Q) using a linear gradient of 0–40% buffer B over 75 ml in a Mono Q column (GE Healthcare) at a flow rate of 0.5 ml min−1. Under these conditions, the enzyme elutes around 15–20% NaCl. Mono Q Buffer A is 50 mM Tris-HCl, pH 8.0. Mono Q Buffer B is 50 mM Tris-HCl (pH 8.0) and 1 M NaCl.
-
16
Add 10% (vol/vol) glycerol to the preparation and divide it into aliquots.
Pause point
Snap-freeze and store the aliquots at −80 °C, where they are stable for a long time (months). Once thawed, sortase A can be stored at 4 °C for 1 month with no noticeable loss of activity.
Reaction setup
Timing 1 d
-
17
Mix a solution containing 10–50 μM target protein, 20–150 μM sortase and 1–2 mM oligoglycine probe in 1× sortase buffer (final concentrations). The controls to be included are as follows: target protein only, sortase only, oligoglycine probe only, target protein and sortase, target protein and oligoglycine probe, sortase and oligoglycine probe.
-
18
Incubate the reactions at RT or at 37 °C. Take 1-μl aliquots after 2, 4, 6, 8 and 16 h. Add 1× LDS gel-loading buffer to the aliquots to stop the reaction and boil it for 2 min.
Pause point
The aliquots can be frozen at −20 °C until further analysis.
-
19
Analyze the aliquots by SDS-PAGE followed by Coomassie blue staining.
Purification of the labeled product
Timing 1 d
-
20
Pipette 500 μl of the Ni-NTA resin (50% slurry) into a microcentrifuge tube. Centrifuge it at 280g for 5 min at 4 °C. Discard the supernatant. Wash the resin three times using 1 ml of nickel-binding buffer.
Critical Step
Do not let the beads dry, as drying decreases their performance.
-
21
Dilute the sortase reaction with nickel-binding buffer to a final volume of 500 μl. (We recommend taking an aliquot of 5 μl from the diluted reaction to later determine the efficiency of purification). Add this to the Ni-NTA resin. Agitate the mixture on an end-over-end shaker for 30 min at 4 °C. These conditions work in most cases. However, keep in mind that the amount of resin to be used per sample depends on the amount of protein that remains unmodified and on the amount of sortase used in the reaction. The protein capacity of the Ni-NTA is ∼50 mg ml−1.
-
22
Centrifuge the mixture at 280g for 5 min at 4 °C. Retain the supernatant.
-
23
Add 500 μl of nickel-binding buffer to the resin and agitate it on an end-over-end shaker for 10 min at 4 °C.
-
24
Centrifuge the mixture at 280g for 5 min at 4 °C. Retain the supernatant.
-
25
Combine the supernatants obtained in Steps 22 and 24. Take an aliquot and analyze it together with the aliquot taken in Step 21 by SDS-PAGE.
-
26
If further purification is required to remove free peptide nucleophile, we recommend using a desalting column equilibrated in a buffer of choice, depending on the use of and compatibility with the labeled protein. Purification by gel filtration (Step 13) is an alternative.
Pause point
The storage conditions depend on the stability of the protein of interest. Most proteins are stable at –70 °C for up to 1 year.
Troubleshooting
Troubleshooting advice can be found in Table 1.
Timing
Step 1A, synthesis of GGGK-biotin and GGGK-TAMRA peptides: 2–3 d
Step 1B, synthesis of GGGK–NHS ester probes: 3 d
Step 1C, synthesis of GGGC–maleimide-containing probes: 3 d
Steps 2–6, transformation of plasmid, expression of sortase A and collection of bacteria: 3 d
Steps 7–16, purification and storage of sortase A: 1–2 d
Steps 17–19, setting up the reaction conditions for sortase-mediated transacylation: 1 d
Steps 20–26, purification of the labeled product: 1 d
Box 1, monitoring of peptide coupling: variable; <1 h
Anticipated results
A typical result of a sortase-based reaction is shown in Figure 4. A successful sortase reaction often results in the formation of the acyl-enzyme intermediate if the oligoglycine probe is omitted. Small quantities of hydrolysis product (loss of His6 or epitope tag) may occur upon cleavage of the sortase motif in the absence of added nucleophile. The acyl-enzyme intermediate usually survives reducing SDS-PAGE in detectable amounts. A full sortase reaction often yields a reaction product of mobility distinct from that of the input substrate and the hydrolysis product. The ability to distinguish the various intermediates critically depends on the molecular weight of the anticipated products and the gel systems used to analyze them. For most proteins, >95% and >50% labeling efficiency is achieved when using a peptide or polypeptide-based nucleophile, respectively. The exact reaction conditions for protein labeling need to be determined empirically for each protein substrate. The range provided has yielded acceptable results in the majority of cases. To achieve maximal levels of labeling, it is helpful to titrate the sortase, substrate and probe concentrations relative to one another, as well as to vary the time of labeling and the temperature of the reaction.
The sortaggable holotoxin was incubated with trypsin to expose the LPETG motif installed in the CTA1 subunit of cholera toxin. Upon incubation with sortase and the nucleophile ubiquitin (comprising a single glycine residue at its N terminus (GUbq)), we infer that most of the CTA1(>90%) is converted to CTA1-GUbq. In our hands, these yields are typical for sortase-mediated transacylations. Note that CTA1-GUbq is detected only when the three reaction components are mixed. The acyl-intermediate conjugate and the product of CTA1 hydrolysis are also highlighted. In addition, although the subunit B of cholera toxin (CTB) is visible, only the subunit CTA1 participates in the reaction. This attests to the specificity of the sortase-labeling reaction even in protein mixtures. A reducing SDS-PAGE gel stained with Coomassie blue is shown. Lane MW shows molecular-weight markers. See ref. 1.
References
Guimaraes, C.P. et al. Identification of host cell factors required for intoxication through use of modified cholera toxin. J. Cell Biol. 195, 751–764 (2011).
Olvera-Gomez, I. et al. Cholera toxin activates nonconventional adjuvant pathways that induce protective CD8 T cell responses after epicutaneous vaccination. Proc. Natl. Acad. Sci. USA 109, 2072–2077 (2012).
Popp, M.W., Karssemeijer, R.A. & Ploegh, H.L. Chemoenzymatic site-specific labeling of influenza glycoproteins as a tool to observe virus budding in real time. PLoS Pathog. 8, e1002604 (2012).
Swee, L.K. et al. Sortase-mediated modification of αDEC205 affords optimization of antigen presentation and immunization against a set of viral epitopes. Proc. Natl. Acad. Sci. USA 110, 1428–1433 (2013).
Muralidharan, V. & Muir, T.W. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat. Methods 3, 429–438 (2006).
Vila-Perello, M. et al. Streamlined expressed protein ligation using split inteins. J. Am. Chem. Soc. 135, 286–292 (2013).
Gautier, A. et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 (2008).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003).
Los, G.V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008).
Howarth, M. & Ting, A.Y. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat. Protoc. 3, 534–545 (2008).
Cohen, J.D., Zou, P. & Ting, A.Y. Site-specific protein modification using lipoic acid ligase and bis-aryl hydrazone formation. Chembiochem 13, 888–894 (2012).
Carrico, I.S., Carlson, B.L. & Bertozzi, C.R. Introducing genetically encoded aldehydes into proteins. Nat. Chem. Biol. 3, 321–322 (2007).
Rabuka, D., Rush, J.S., deHart, G.W., Wu, P. & Bertozzi, C.R. Site-specific chemical protein conjugation using genetically encoded aldehyde tags. Nat. Protoc. 7, 1052–1067 (2012).
Yin, J., Lin, A.J., Golan, D.E. & Walsh, C.T. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 1, 280–285 (2006).
Parthasarathy, R., Subramanian, S. & Boder, E.T. Sortase A as a novel molecular 'stapler' for sequence-specific protein conjugation. Bioconjug. Chem. 18, 469–476 (2007).
Popp, M.W., Antos, J.M., Grotenbreg, G.M., Spooner, E. & Ploegh, H.L. Sortagging: a versatile method for protein labeling. Nat. Chem. Biol. 3, 707–708 (2007).
Popp, M.W. & Ploegh, H.L. Making and breaking peptide bonds: protein engineering using sortase. Angew. Chem. Int. Ed. Engl. 50, 5024–5032 (2011).
Ton-That, H., Mazmanian, S.K., Faull, K.F. & Schneewind, O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J. Biol. Chem. 275, 9876–9881 (2000).
Theile, C.S. et al. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat. Protoc. 8, 1800–1807 (2013).
Witte, M.D. et al. Production of unnaturally linked chimeric proteins using a combination of sortase-catalyzed transpeptidation and click chemistry. Nat. Protoc. 8, 1808–1819 (2013).
Marraffini, L.A., Dedent, A.C. & Schneewind, O. Sortases and the art of anchoring proteins to the envelopes of Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70, 192–221 (2006).
Mazmanian, S.K., Liu, G., Ton-That, H. & Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 (1999).
Spirig, T., Weiner, E.M. & Clubb, R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 82, 1044–1059 (2011).
Ton-That, H., Liu, G., Mazmanian, S.K., Faull, K.F. & Schneewind, O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96, 12424–12429 (1999).
Antos, J.M., Miller, G.M., Grotenbreg, G.M. & Ploegh, H.L. Lipid modification of proteins through sortase-catalyzed transpeptidation. J. Am. Chem. Soc. 130, 16338–16343 (2008).
Pritz, S. et al. Synthesis of biologically active peptide nucleic acid-peptide conjugates by sortase-mediated ligation. J. Org. Chem. 72, 3909–3912 (2007).
Samantaray, S., Marathe, U., Dasgupta, S., Nandicoori, V.K. & Roy, R.P. Peptide-sugar ligation catalyzed by transpeptidase sortase: a facile approach to neoglycoconjugate synthesis. J. Am. Chem. Soc. 130, 2132–2133 (2008).
Sinisi, A. et al. Development of an influenza virus protein array using sortagging technology. Bioconjug. Chem. 23, 1119–1126 (2012).
Witte, M.D. et al. Preparation of unnatural N-to-N and C-to-C protein fusions. Proc. Natl. Acad. Sci. USA 109, 11993–11998 (2012).
Antos, J.M. et al. A straight path to circular proteins. J. Biol. Chem. 284, 16028–16036 (2009).
Williamson, D.J., Fascione, M.A., Webb, M.E. & Turnbull, W.B. Efficient N-terminal labeling of proteins by use of sortase. Angew. Chem. Int. Ed. Engl. 51, 9377–9380 (2012).
Levary, D.A., Parthasarathy, R., Boder, E.T. & Ackerman, M.E. Protein-protein fusion catalyzed by sortase A. PLoS ONE 6, e18342 (2011).
Popp, M.W., Dougan, S.K., Chuang, T.Y., Spooner, E. & Ploegh, H.L. Sortase-catalyzed transformations that improve the properties of cytokines. Proc. Natl. Acad. Sci. USA 108, 3169–3174 (2011).
Popp, M.W., Artavanis-Tsakonas, K. & Ploegh, H.L. Substrate filtering by the active site crossover loop in UCHL3 revealed by sortagging and gain-of-function mutations. J. Biol. Chem. 284, 3593–3602 (2009).
Bolscher, J.G. et al. Sortase A as a tool for high-yield histatin cyclization. FASEB J. 25, 2650–2658 (2011).
Wu, Z., Guo, X. & Guo, Z. Sortase A-catalyzed peptide cyclization for the synthesis of macrocyclic peptides and glycopeptides. Chem. Commun. 47, 9218–9220 (2011).
Chen, I., Dorr, B.M. & Liu, D.R. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. USA 108, 11399–11404 (2011).
Hirakawa, H., Ishikawa, S. & Nagamune, T. Design of Ca2+-independent Staphylococcus aureus sortase A mutants. Biotechnol. Bioeng. 109, 2955–2961 (2012).
Race, P.R. et al. Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J. Biol. Chem. 284, 6924–6933 (2009).
Antos, J.M. et al. Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. J. Am. Chem. Soc. 131, 10800–10801 (2009).
Esteban, A. et al. Fungal recognition is mediated by the association of dectin-1 and galectin-3 in macrophages. Proc. Natl. Acad. Sci. USA 108, 14270–14275 (2011).
Hess, G.T. et al. M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins. Bioconjug. Chem. 23, 1478–1487 (2012).
Steinhagen, M., Zunker, K., Nordsieck, K. & Beck-Sickinger, A.G. Large scale modification of biomolecules using immobilized sortase A from Staphylococcus aureus. Bioorg. Med. Chem. 21, 3504–3510 (2013).
Ilangovan, U., Ton-That, H., Iwahara, J., Schneewind, O. & Clubb, R.T. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98, 6056–6061 (2001).
Boekhorst, J., de Been, M.W., Kleerebezem, M. & Siezen, R.J. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J. Bacteriol. 187, 4928–4934 (2005).
Kaiser, E., Colescott, R.L., Bossinger, C.D. & Cook, P.I. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 34, 595–598 (1970).
Gisin, B.F. The monitoring of reactions in solid-phase peptide synthesis with picric acid. Anal. Chim. Acta. 58, 248–249 (1972).
Vojkovsky, T. Detection of secondary amines on solid phase. Pept. Res. 8, 236–237 (1995).
Acknowledgements
This work was supported by funding from The Netherlands Organisation for Scientific Research (to M.D.W.) and the US National Institutes of Health (NIH; grant no. RO1 AI087879 to H.L.P.).
Author information
Authors and Affiliations
Contributions
C.P.G. and H.L.P. conceived of and drafted the manuscript; C.P.G., M.D.W., C.S.T., L.K. and G.B. participated in the optimization of the protocols and wrote the manuscript. A.E.M.B. and H.L.P. revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Guimaraes, C., Witte, M., Theile, C. et al. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat Protoc 8, 1787–1799 (2013). https://doi.org/10.1038/nprot.2013.101
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.101
This article is cited by
-
Dynamic stability of Sgt2 enables selective and privileged client handover in a chaperone triad
Nature Communications (2024)
-
Assembly mechanism of the inflammasome sensor AIM2 revealed by single molecule analysis
Nature Communications (2023)
-
Turning antibodies off and on again using a covalently tethered blocking peptide
Communications Biology (2022)
-
Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation
Nature Communications (2022)
-
Preclinical evaluation of [99mTc]Tc-labeled anti-EpCAM nanobody for EpCAM receptor expression imaging by immuno-SPECT/CT
European Journal of Nuclear Medicine and Molecular Imaging (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.