Abstract
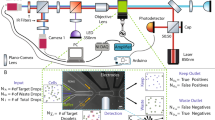
We present a droplet-based microfluidics protocol for high-throughput analysis and sorting of single cells. Compartmentalization of single cells in droplets enables the analysis of proteins released from or secreted by cells, thereby overcoming one of the major limitations of traditional flow cytometry and fluorescence-activated cell sorting. As an example of this approach, we detail a binding assay for detecting antibodies secreted from single mouse hybridoma cells. Secreted antibodies are detected after only 15 min by co-compartmentalizing single mouse hybridoma cells, a fluorescent probe and single beads coated with anti-mouse IgG antibodies in 50-pl droplets. The beads capture the secreted antibodies and, when the captured antibodies bind to the probe, the fluorescence becomes localized on the beads, generating a clearly distinguishable fluorescence signal that enables droplet sorting at ∼200 Hz as well as cell enrichment. The microfluidic system described is easily adapted for screening other intracellular, cell-surface or secreted proteins and for quantifying catalytic or regulatory activities. In order to screen ∼1 million cells, the microfluidic operations require 2–6 h; the entire process, including preparation of microfluidic devices and mammalian cells, requires 5–7 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Guo, M.T., Rotem, A., Heyman, J.A. & Weitz, D.A. Droplet microfluidics for high-throughput biological assays. Lab Chip 12, 2146–2155 (2012).
Kintses, B., van Vliet, L.D., Devenish, S.R. & Hollfelder, F. Microfluidic droplets: new integrated workflows for biological experiments. Curr. Opin. Chem. Biol. 14, 548–555 (2010).
Theberge, A.B. et al. Microdroplets in microfluidics: an evolving platform for discoveries in chemistry and biology. Angew. Chem. Int. Ed. 49, 5846–5868 (2010).
Dove, A. Drug screening--beyond the bottleneck. Nat. Biotechnol. 17, 859–863 (1999).
Link, D.R., Anna, S.L., Weitz, D.A. & Stone, H.A. Geometrically mediated breakup of drops in microfluidic devices. Phys. Rev. Lett. 92, 054503 (2004).
Mazutis, L., Baret, J.C. & Griffiths, A.D. A fast and efficient microfluidic system for highly selective one-to-one droplet fusion. Lab Chip 9, 2665–2672 (2009).
Mazutis, L. & Griffiths, A.D. Selective droplet coalescence using microfluidic systems. Lab Chip 12, 1800–1806 (2012).
Ahn, K., Agresti, J., Chong, H., Marquez, M. & Weitz, D.A. Electrocoalescence of drops synchronized by size-dependent flow in microfluidic channels. Appl. Phys. Lett. 88, 264105–264103 (2006).
Chabert, M., Dorfman, K.D. & Viovy, J.L. Droplet fusion by alternating current (AC) field electrocoalescence in microchannels. Electrophoresis 26, 3706–3715 (2005).
Priest, C., Herminghaus, S. & Seemann, R. Controlled electrocoalescence in microfluidics: targeting a single lamella. Appl. Phys. Lett. 89, 134101 (2006).
Abate, A.R., Hung, T., Mary, P., Agresti, J.J. & Weitz, D.A. High-throughput injection with microfluidics using picoinjectors. Proc. Natl. Acad. Sci. USA 107, 19163–19166 (2010).
Li, L. et al. Nanoliter microfluidic hybrid method for simultaneous screening and optimization validated with crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA 103, 19243–19248 (2006).
Niu, X., Gulati, S., Edel, J.B. & deMello, A.J. Pillar-induced droplet merging in microfluidic circuits. Lab Chip 8, 1837–1841 (2008).
Frenz, L., Blank, K., Brouzes, E. & Griffiths, A.D. Reliable microfluidic on-chip incubation of droplets in delay-lines. Lab Chip 9, 1344–1348 (2009).
Hatch, A.C. et al. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip 11, 3838–3845 (2011).
Shim, J.U. et al. Simultaneous determination of gene expression and enzymatic activity in individual bacterial cells in microdroplet compartments. J. Am. Chem. Soc. 42, 15251–15256 (2009).
Mazutis, L. et al. Droplet-based microfluidic systems for high-throughput single DNA molecule isothermal amplification and analysis. Anal. Chem. 81, 4813–4821 (2009).
Lichtman, J.W. & Conchello, J.A. Fluorescence microscopy. Nat. Methods 2, 910–919 (2005).
Najah, M., Griffiths, A.D. & Ryckelynck, M. Teaching single-cell digital analysis using droplet-based microfluidics. Anal. Chem. 84, 1202–1209 (2012).
Ahn, K. et al. Dielectrophoretic manipulation of drops for high-speed microfluidic sorting devices. Appl. Phys. Lett. 88, 024104 (2006).
Franke, T., Abate, A.R., Weitz, D.A. & Wixforth, A. Surface acoustic wave (SAW) directed droplet flow in microfluidics for PDMS devices. Lab Chip 9, 2625–2627 (2009).
Debs, B.E., Utharala, R., Balyasnikova, I.V., Griffiths, A.D. & Merten, C.A. Functional single-cell hybridoma screening using droplet-based microfluidics. Proc. Natl. Acad. Sci. USA 109, 11570–11575 (2012).
Granieri, L., Baret, J.C., Griffiths, A.D. & Merten, C.A. High-throughput screening of enzymes by retroviral display using droplet-based microfluidics. Chem. Biol. 17, 229–235 (2010).
He, M. et al. Selective encapsulation of single cells and subcellular organelles into picoliter- and femtoliter-volume droplets. Anal. Chem. 77, 1539–1544 (2005).
Clausell-Tormos, J. et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem. Biol. 15, 427–437 (2008).
Koster, S. et al. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip 8, 1110–1115 (2008).
Brouzes, E. et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 106, 14195–14200 (2009).
Liu, W., Kim, H.J., Lucchetta, E.M., Du, W. & Ismagilov, R.F. Isolation, incubation, and parallel functional testing and identification by FISH of rare microbial single-copy cells from multi-species mixtures using the combination of chemistrode and stochastic confinement. Lab Chip 9, 2153–2162 (2009).
Hufnagel, H. et al. An integrated cell culture lab on a chip: modular microdevices for cultivation of mammalian cells and delivery into microfluidic microdroplets. Lab Chip 9, 1576–1582 (2009).
Zeng, Y., Novak, R., Shuga, J., Smith, M.T. & Mathies, R.A. High-performance single cell genetic analysis using microfluidic emulsion generator arrays. Anal. Chem. 82, 3183–3190 (2010).
Rane, T.D., Zec, H.C., Puleo, C., Lee, A.P. & Wang, T.H. Droplet microfluidics for amplification-free genetic detection of single cells. Lab Chip 12, 3341–3347 (2012).
Huebner, A. et al. Development of quantitative cell-based enzyme assays in microdroplets. Anal. Chem. 80, 3890–3896 (2008).
Baret, J.C., Beck, Y., Billas-Massobrio, I., Moras, D. & Griffiths, A.D. Quantitative cell-based reporter gene assays using droplet-based microfluidics. Chem. Biol. 17, 528–536 (2010).
Huebner, A. et al. Quantitative detection of protein expression in single cells using droplet microfluidics. Chem. Commun. (Camb) 28, 1218–1220 (2007).
Chen, D. et al. The chemistrode: a droplet-based microfluidic device for stimulation and recording with high temporal, spatial, and chemical resolution. Proc. Natl. Acad. Sci. USA 105, 16843–16848 (2008).
Niu, X., Gielen, F., Edel, J.B. & deMello, A.J. A microdroplet dilutor for high-throughput screening. Nat. Chem. 3, 437–442 (2011).
Mazutis, L. et al. Multi-step microfluidic droplet processing: kinetic analysis of an in vitro–translated enzyme. Lab Chip 9, 2902–2908 (2009).
Pekin, D. et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip 11, 2156–2166 (2011).
Zhong, Q. et al. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab Chip 11, 2167–2174 (2011).
Hindson, B.J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610 (2011).
Tewhey, R. et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat. Biotechnol. 27, 1025–1031 (2009).
Miller, O.J. et al. High-resolution dose-response screening using droplet-based microfluidics. Proc. Natl. Acad. Sci. USA 109, 378–383 (2012).
Clausell-Tormos, J., Griffiths, A.D. & Merten, C.A. An automated two-phase microfluidic system for kinetic analyses and the screening of compound libraries. Lab Chip 10, 1302–1307 (2010).
Churski, K. et al. Rapid screening of antibiotic toxicity in an automated microdroplet system. Lab Chip 12, 1629–1637 (2012).
Agresti, J.J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 107, 4004–4009 (2010).
Kintses, B. et al. Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem. Biol. 19, 1001–1009 (2012).
Paegel, B.M. & Joyce, G.F. Microfluidic compartmentalized directed evolution. Chem. Biol. 17, 717–724 (2010).
Lowe, K.C. Perfluorochemical respiratory gas carriers: benefits to cell culture systems. J. Fluorine Chem. 118, 19–26 (2002).
Scott, R.L. The solubility of fluorocarbons. J. Am. Chem. Soc. 70, 4090–4093 (1948).
Simons, J.H. & Linevsky, M.J. The solubility of organic solids in fluorocarbon derivatives. J. Am. Chem. Soc. 74, 4750–4751 (1952).
Tawfik, D.S. & Griffiths, A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 16, 652–656 (1998).
Baret, J.C. et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip 9, 1850–1858 (2009).
Martino, C. et al. Intracellular protein determination using droplet-based immunoassays. Anal. Chem. 83, 5361–5368 (2011).
Beer, N.R. et al. On-chip single-copy real-time reverse-transcription PCR in isolated picoliter droplets. Anal. Chem. 80, 1854–1858 (2008).
Schaerli, Y. et al. Continuous-flow polymerase chain reaction of single-copy DNA in microfluidic microdroplets. Anal. Chem. 81, 302–306 (2009).
Beer, N.R. et al. On-chip, real-time, single-copy polymerase chain reaction in picoliter droplets. Anal. Chem. 79, 8471–8475 (2007).
Olsen, M.J. et al. Function-based isolation of novel enzymes from a large library. Nat. Biotechnol. 18, 1071–1074 (2000).
Aharoni, A. et al. High-throughput screening methodology for the directed evolution of glycosyltransferases. Nat. Methods 3, 609–614 (2006).
Unger, M.A., Chou, H.P., Thorsen, T., Scherer, A. & Quake, S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116 (2000).
Weinstein, J.A., Jiang, N., White, R.A. III, Fisher, D.S. & Quake, S.R. High-throughput sequencing of the zebrafish antibody repertoire. Science 324, 807–810 (2009).
Fan, H.C., Wang, J., Potanina, A. & Quake, S.R. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 29, 51–57 (2011).
Lecault, V. et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat. Methods 8, 581–586 (2011).
Love, J.C., Ronan, J.L., Grotenbreg, G.M., van der Veen, A.G. & Ploegh, H.L. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat. Biotechnol. 24, 703–707 (2006).
Taly, V., Pekin, D., El Abed, A. & Laurent-Puig, P. Detecting biomarkers with microdroplet technology. Trends Mol. Med. 18, 405–416 (2012).
Choi, J.W., Kang, D.K., Park, H., deMello, A.J. & Chang, S.I. High-throughput analysis of protein-protein interactions in picoliter-volume droplets using fluorescence polarization. Anal. Chem. 84, 3849–3854 (2012).
Joensson, H.N., Zhang, C., Uhlen, M. & Andersson-Svahn, H. A homogeneous assay for protein analysis in droplets by fluorescence polarization. Electrophoresis 33, 436–439 (2012).
Srisa-Art, M., Dyson, E.C., Demello, A.J. & Edel, J.B. Monitoring of real-time streptavidin-biotin binding kinetics using droplet microfluidics. Anal. Chem. 80, 7063–7067 (2008).
Cecchini, M.P. et al. Ultrafast surface enhanced resonance Raman scattering detection in droplet-based microfluidic systems. Anal. Chem. 83, 3076–3081 (2011).
Reymond, J.L., Fluxa, V.S. & Maillard, N. Enzyme assays. Chem. Commun. 2009, 34–46 (2009).
Joensson, H.N. et al. Detection and analysis of low-abundance cell-surface biomarkers using enzymatic amplification in microfluidic droplets. Angew. Chem. Int. Ed. Engl. 48, 2518–2521 (2009).
Xia, Y.N. & Whitesides, G.M. Soft lithography. Angew. Chem. Int. Ed. 37, 551–575 (1998).
Anna, S.L., Bontoux, N. & Stone, H.A. Formation of dispersions using “flow focusing” in microchannels. Appl. Phys. Lett. 82, 364–366 (2003).
Garstecki, P., Fuerstman, M.J., Stone, H.A. & Whitesides, G.M. Formation of droplets and bubbles in a microfluidic T-junction—scaling and mechanism of break-up. Lab Chip 6, 437–446 (2006).
Abate, A.R. et al. Impact of inlet channel geometry on microfluidic drop formation. Phys. Rev. E 80, 026310 (2009).
Sugiura, S., Nakajima, M., Iwamoto, S. & Seki, M. Interfacial tension driven monodispersed droplet formation from microfabricated channel array. Langmuir 17, 5562–5566 (2001).
Baret, J.C. Surfactants in droplet-based microfluidics. Lab Chip 12, 422–433 (2012).
Baret, J.C., Kleinschmidt, F., El Harrak, A. & Griffiths, A.D. Kinetic aspects of emulsion stabilization by surfactants: a microfluidic analysis. Langmuir 25, 6088–6093 (2009).
Roach, L.S., Song, H. & Ismagilov, R.F. Controlling nonspecific protein adsorption in a plug-based microfluidic system by controlling interfacial chemistry using fluorous-phase surfactants. Anal. Chem. 77, 785–796 (2005).
Holtze, C. et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 8, 1632–1639 (2008).
Chen, F. et al. Chemical transfection of cells in picoliter aqueous droplets in fluorocarbon oil. Anal. Chem. 83, 8816–8820 (2011).
Hudlicky, M. & Pavlath, A.E. Chemistry of Organic Fluorine Compounds. (American Chemical Society, 1995).
Skhiri, Y. Dynamics of molecular transport by surfactants in emulsions. Soft Matter 8, 10618–10627 (2012).
Woronoff, G. et al. New generation of amino coumarin methyl sulfonate-based fluorogenic substrates for amidase assays in droplet-based microfluidic applications. Anal. Chem. 83, 2852–2857 (2011).
Courtois, F. et al. Controlling the retention of small molecules in emulsion microdroplets for use in cell-based assays. Anal. Chem. 81, 3008–3016 (2009).
Siegel, A.C. et al. Cofabrication of electromagnets and microfluidic systems in poly(dimethylsiloxane). Angew Chem. Int. Ed. Engl. 45, 6877–6882 (2006).
Asmolov, E.S. The inertial lift on a spherical particle in a plane Poiseuille flow at large channel Reynolds number. J. Fluid Mech. 381, 63–87 (1999).
Di Carlo, D., Irimia, D., Tompkins, R.G. & Toner, M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc. Natl. Acad. Sci. USA 104, 18892–18897 (2007).
Kemna, E.W.M. et al. High-yield cell ordering and deterministic cell-in-droplet encapsulation using Dean flow in a curved microchannel. Lab Chip 12, 2881–2887 (2012).
Ford, T., Graham, J. & Rickwood, D. Iodixanol—a nonionic isosmotic centrifugation medium for the formation of self-generated gradients. Anal. Biochem. 220, 360–366 (1994).
Bruce, A.T. et al. Use of iodixanol self-generated density gradients to enrich for viable urothelial cells from nonneurogenic and neurogenic bladder tissue. Tissue Eng. Part C Methods 16, 33–40 (2010).
Graziani-Bowering, G.M., Graham, J.M. & Filion, L.G. A quick, easy and inexpensive method for the isolation of human peripheral blood monocytes. J. Immunol. Methods 207, 157–168 (1997).
Burgoyne, F. A remote syringe for cells, beads and particle injection in microfluidic channels. http://blogs.rsc.org/chipsandtips/2009/08/20/a-remote-syringe-for-cells-beads-and-particle-injection-in-microfluidic-channels/ (20 August 2009).
Shapiro, H.M. Practical Flow Cytometry. 4th edn. (Wiley-Liss, 2003).
Moon, S., Ceyhan, E., Gurkan, U.A. & Demirci, U. Statistical modeling of single target cell encapsulation. PLoS ONE 6, e21580 (2011).
Chabert, M. & Viovy, J.L. Microfluidic high-throughput encapsulation and hydrodynamic self-sorting of single cells. Proc. Natl. Acad. Sci. USA 105, 3191–3196 (2008).
Acknowledgements
We are grateful to R. Sperling for the kind gift of surfactant and D. Aubrecht for his help in developing droplet detection and sorting methods. This work was supported by the National Science Foundation (NSF) (DMR-1006546), the National Institutes of Health (NIH) (P01GM096971 and 5R01EB014703-02), the Harvard Materials Science Research and Engineering Center (DMR-0820484) and the Lithuanian Research Council (MIP-048/2012). W.L.U. acknowledges support from a Canadian National Sciences and Engineering Research Council (NSERC) Postgraduate Scholarship (PGS D). J.A.H. and J.G. were supported by NIH SBIR grant no. 1R43AI082861-01. This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the NSF under award no. ECS-0335765. CNS is part of Harvard University.
Author information
Authors and Affiliations
Contributions
L.M. and J.A.H. performed the experiments described in this protocol, W.L.U. provided the LabVIEW software, L.M., J.A.H. and A.D.G. analyzed the data; J.G. set up the detection system, all authors edited and proofread the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors are inventors on a patent application (PCT/US2008/008563) including some of the ideas described in this manuscript.
Supplementary information
Supplementary Video 1
Cell and bead co-encapsulation (MOV 353 kb)
Supplementary Video 2
Droplet containing cells and beads reinjection and spacing (MOV 1756 kb)
Supplementary Video 3
Droplet sorting (MOV 495 kb)
Supplementary Data (*dwg format, to be viewed with AutoCAD software)
AutoCad designs of the microfluidic chips (ZIP 1189 kb)
Supplementary Figure 1
Droplet volume as a function of flow rate. Microfluidic channels were 25 μm deep and the design is provided in Supplementary Material and indicated in Figure 2a. The flow rate for the aqueous phase was kept at 180 μl/h, while the flow rate for the continuous phase was varied from 160 μl/h to 300 μl/h (PDF 285 kb)
Supplementary Note
Plate-based sandwich assay to measure secreted antibody (PDF 258 kb)
Rights and permissions
About this article
Cite this article
Mazutis, L., Gilbert, J., Ung, W. et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc 8, 870–891 (2013). https://doi.org/10.1038/nprot.2013.046
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.046
This article is cited by
-
Design automation of microfluidic single and double emulsion droplets with machine learning
Nature Communications (2024)
-
Small-molecule autocatalysis drives compartment growth, competition and reproduction
Nature Chemistry (2024)
-
Harnessing 3D in vitro systems to model immune responses to solid tumours: a step towards improving and creating personalized immunotherapies
Nature Reviews Immunology (2024)
-
A mini review on recent progress of microfluidic systems for antibody development
Journal of Diabetes & Metabolic Disorders (2024)
-
A home-made pipette droplet microfluidics rapid prototyping and training kit for digital PCR, microorganism/cell encapsulation and controlled microgel synthesis
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.