Abstract
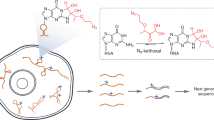
RNA structure is important for RNA function and regulation, and there is growing interest in determining the RNA structure of many transcripts. Here we provide a detailed protocol for the parallel analysis of RNA structure (PARS) for probing RNA secondary structures genome-wide. In this method, enzymatic footprinting is coupled to high-throughput sequencing to provide secondary structure data for thousands of RNAs simultaneously. The entire experimental protocol takes ∼5 d to complete, and sequencing and data analysis take an additional 6–8 d. PARS was developed using the yeast genome as proof of principle, but its approach should be applicable to probing RNA structures from different transcriptomes and structural dynamics under diverse solution conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sharp, P.A. The centrality of RNA. Cell 136, 577–580 (2009).
Wan, Y., Kertesz, M., Spitale, R.C., Segal, E. & Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 12, 641–655 (2011).
Breaker, R.R. Prospects for riboswitch discovery and analysis. Mol. Cell 43, 867–879 (2011).
Guo, F., Gooding, A.R. & Cech, T.R. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol. Cell 16, 351–362 (2004).
Deigan, K.E., Li, T.W., Mathews, D.H. & Weeks, K.M. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA 106, 97–102 (2009).
Gornicki, P. et al. Use of lead(II) to probe the structure of large RNA's. Conformation of the 3′ terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J. Biomol. Struct. Dyn. 6, 971–984 (1989).
Auron, P.E., Weber, L.D. & Rich, A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry 21, 4700–4706 (1982).
Watts, J.M. et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716 (2009).
Wilkinson, K.A. et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6, e96 (2008).
Kertesz, M. et al. Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107 (2010).
Zheng, Q. et al. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 6, e1001141 (2010).
Underwood, J.G. et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods 7, 995–1001 (2010).
Lucks, J.B. et al. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. USA 108, 11063–11068 (2011).
Li, F. et al. Global analysis of RNA secondary structure in two metazoans. Cell Rep. 1, 69–82 (2012).
Ouyang, Z., Snyder, M.P. & Chang, H.Y. SeqFold: Genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data. Genome Res. (2012).
Wan, Y. et al. Genome-wide measurement of RNA folding energies. Mol. Cell 48, 169–181 (2012).
Merino, E.J., Wilkinson, K.A., Coughlan, J.L. & Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 (2005).
Low, J.T. & Weeks, K.M. SHAPE-directed RNA secondary structure prediction. Methods 52, 150–158 (2010).
Wurst, R.M., Vournakis, J.N. & Maxam, A.M. Structure mapping of 5′-32P-labeled RNA with S1 nuclease. Biochemistry 17, 4493–4499 (1978).
Lowman, H.B. & Draper, D.E. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 261, 5396–5403 (1986).
Ehresmann, C. et al. Probing the structure of RNAs in solution. Nucleic Acids Res. 15, 9109–9128 (1987).
Lee, A., Hansen, K.D., Bullard, J., Dudoit, S. & Sherlock, G. Novel low abundance and transient RNAs in yeast revealed by tiling microarrays and ultra high-throughput sequencing are not conserved across closely related yeast species. PLoS Genet. 4, e1000299 (2008).
Neil, H. et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457, 1038–1042 (2009).
Xu, Z. et al. Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037 (2009).
ENCODE Project Consortium et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Rumble, S.M. et al. SHRiMP: accurate mapping of short color-space reads. PLoS Comput. Biol. 5, e1000386 (2009).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 (2008).
Ding, Y., Chan, C.Y. & Lawrence, C.E. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 11, 1157–1166 (2005).
Saldanha, A.J. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004).
Darty, K., Denise, A. & Ponty, Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 (2009).
Quail, M.A. et al. A large genome center's improvements to the Illumina sequencing system. Nat. Methods 5, 1005–1010 (2008).
Zwieb, C., van Nues, R.W., Rosenblad, M.A., Brown, J.D. & Samuelsson, T.A nomenclature for all signal recognition particle RNAs. RNA 11, 7–13 (2005).
Acknowledgements
This work was supported by National Institutes of Health (R01-HG004361). Y.W. is funded by the Agency of Science, Technology and Research of Singapore. H.Y.C. is an Early Career Scientist of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
Y.W. and H.Y.C. developed the protocol and designed the experiments; Y.W. performed the experiments; K.Q. analyzed the data; Z.O. developed the SeqFold pipeline; Y.W. and H.Y.C. wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
Y.W. and H.Y.C. are named as inventors on a patent application filed on the PARS method by Weizmann Institute and Stanford University.
Rights and permissions
About this article
Cite this article
Wan, Y., Qu, K., Ouyang, Z. et al. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat Protoc 8, 849–869 (2013). https://doi.org/10.1038/nprot.2013.045
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.045
This article is cited by
-
Getting to the bottom of lncRNA mechanism: structure–function relationships
Mammalian Genome (2022)
-
Systematic evaluation and optimization of the experimental steps in RNA G-quadruplex structure sequencing
Scientific Reports (2019)
-
Comparative and integrative analysis of RNA structural profiling data: current practices and emerging questions
Quantitative Biology (2017)
-
The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA
Nature (2016)
-
Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE
Nature Protocols (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.