Abstract
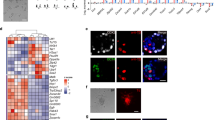
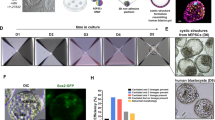
Little is known about the true developmental origin of human embryonic stem cells (hESCs) or the events that initiate their generation. Recently, we have shown that hESCs originate from a post–inner cell mass (ICM) intermediate (PICMI), a unique transient epiblast-like structure that is different from both its ICM progenitor and its subsequent hESC fate. As a closer progenitor of hESCs than the ICM, the PICMI could be used to provide further insight into the human pluripotent state. Here we provide a detailed (7-d) protocol for the culture of the human preimplantation embryos in order to derive the PICMI. Subsequent identification and cryopreservation of the PICMI are described, in addition to hESC derivation. The initial hESC outgrowth is visible within 2–7 d after PICMI plating. By using the protocol provided, we observed PICMI formation in 21.3% of plated blastocysts with good-quality ICMs. Of the PICMIs used for hESC derivation, 80.6% showed hESC outgrowth after further culture.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brons, I.G. et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 (2007).
Tesar, P.J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007).
Najm, F.J. et al. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell 8, 318–325 (2011).
Guo, G. et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 (2009).
Hanna, J. et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell 4, 513–524 (2009).
O'Leary, T. et al. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat. Biotechnol. 30, 278–282 (2012).
Adewumi, O. et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25, 803–816 (2007).
Blair, K., Wray, J. & Smith, A. The liberation of embryonic stem cells. PLoS Genet. 7, e1002019 (2011).
Hanna, J.H., Saha, K. & Jaenisch, R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143, 508–525 (2010).
Osafune, K. et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26, 313–315 (2008).
Hanna, J. et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl Acad. Sci. USA 107, 9222–9227 (2010).
Nichols, J. & Smith, A. The origin and identity of embryonic stem cells. Development 138, 3–8 (2011).
Bao, S. et al. The germ cell determinant Blimp1 is not required for derivation of pluripotent stem cells. Cell Stem Cell 11, 110–117 (2012).
Chu, L.F., Surani, M.A., Jaenisch, R. & Zwaka, T.P. Blimp1 expression predicts embryonic stem cell development in vitro. Curr. Biol. 21, 1759–1765 (2011).
Zwaka, T.P. & Thomson, J.A. A germ cell origin of embryonic stem cells? Development 132, 227–233 (2005).
Kuijk, E.W., Chuva de Sousa Lopes, S.M., Geijsen, N., Macklon, N. & Roelen, B.A. The different shades of mammalian pluripotent stem cells. Hum. Reprod. Update 17, 254–271 (2011).
Yamagata, K., Ueda, J., Mizutani, E., Saitou, M. & Wakayama, T. Survival and death of epiblast cells during embryonic stem cell derivation revealed by long-term live-cell imaging with an Oct4 reporter system. Dev. Biol. 346, 90–101 (2010).
Chen, H. et al. The derivation of two additional human embryonic stem cell lines from day 3 embryos with low morphological scores. Hum. Reprod. 20, 2201–2206 (2005).
Genbacev, O. et al. Serum-free derivation of human embryonic stem cell lines on human placental fibroblast feeders. Fertil. Steril. 83, 1517–1529 (2005).
Lerou, P.H. et al. Human embryonic stem cell derivation from poor-quality embryos. Nat. Biotechnol. 26, 212–214 (2008).
Lysdahl, H. et al. Derivation and characterization of four new human embryonic stem cell lines: the Danish experience. Reprod. Biomed. Online 12, 119–126 (2006).
O'Leary, T. et al. The influence of early embryo traits on human embryonic stem cell derivation efficiency. Stem Cells Dev. 20, 785–793 (2011).
Hasegawa, K., Pomeroy, J.E. & Pera, M.F. Current technology for the derivation of pluripotent stem cell lines from human embryos. Cell Stem Cell 6, 521–531 (2010).
Solter, D. & Knowles, B.B. Immunosurgery of mouse blastocyst. Proc. Natl Acad. Sci. USA 72, 5099–5102 (1975).
Chen, A.E. et al. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell 4, 103–106 (2009).
Findikli, N., Kahraman, S., Akcin, O., Sertyel, S. & Candan, Z. Establishment and characterization of new human embryonic stem cell lines. Reprod. Biomed. Online 10, 617–627 (2005).
Heins, N. et al. Derivation, characterization, and differentiation of human embryonic stem cells. Stem Cells 22, 367–376 (2004).
Tropel, P. et al. High-efficiency derivation of human embryonic stem cell lines following pre-implantation genetic diagnosis. In Vitro Cell Dev. Biol. Anim. 46, 376–385 (2010).
Strom, S. et al. No relationship between embryo morphology and successful derivation of human embryonic stem cell lines. PLoS ONE 5, e15329 (2010).
Fisch, J.D., Rodriguez, H., Ross, R., Overby, G. & Sher, G. The Graduated Embryo Score (GES) predicts blastocyst formation and pregnancy rate from cleavage-stage embryos. Hum. Reprod. 16, 1970–1975 (2001).
Giorgetti, C. et al. Embryo score to predict implantation after in vitro fertilization: based on 957 single embryo transfers. Hum. Reprod. 10, 2427–2431 (1995).
Holte, J. et al. Construction of an evidence-based integrated morphology cleavage embryo score for implantation potential of embryos scored and transferred on day 2 after oocyte retrieval. Hum. Reprod. 22, 548–557 (2007).
Steer, C.V., Mills, C.L., Tan, S.L., Campbell, S. & Edwards, R.G. The cumulative embryo score: a predictive embryo scoring technique to select the optimal number of embryos to transfer in an in vitro fertilization and embryo transfer programme. Hum. Reprod. 7, 117–119 (1992).
Niakan, K.K., Han, J., Pedersen, R.A., Simon, C. & Pera, R.A. Human pre-implantation embryo development. Development 139, 829–841 (2012).
Ottosen, L.D., Kesmodel, U., Hindkjaer, J. & Ingerslev, H.J. Pregnancy prediction models and eSET criteria for IVF patients—do we need more information? J. Assist. Reprod. Genet. 24, 29–36 (2007).
van Loendersloot, L.L. et al. Predictive factors in in vitro fertilization (IVF): a systematic review and meta-analysis. Hum. Reprod. Update 16, 577–589 (2010).
Broekmans, F.J., Knauff, E.A., te Velde, E.R., Macklon, N.S. & Fauser, B.C. Female reproductive ageing: current knowledge and future trends. Trends Endocrinol. Metab. 18, 58–65 (2007).
O'Leary, T. et al. The influence of patient and cohort parameters on the incidence and developmental potential of embryos with poor quality traits for use in human embryonic stem cell derivation. Hum. Reprod. 27, 1581–1589 (2012).
Banerjee, P. et al. Deep phenotyping to predict live birth outcomes in in vitro fertilization. Proc. Natl Acad. Sci. USA 107, 13570–13575 (2010).
Cai, Q.F., Wan, F., Huang, R. & Zhang, H.W. Factors predicting the cumulative outcome of IVF/ICSI treatment: a multivariable analysis of 2450 patients. Hum. Reprod. 26, 2532–2540 (2011).
Jun, S.H. et al. Defining human embryo phenotypes by cohort-specific prognostic factors. PLoS One 3, e2562 (2008).
Inamdar, M.S., Venu, P., Srinivas, M.S., Rao, K. & VijayRaghavan, K. Derivation and characterization of two sibling human embryonic stem cell lines from discarded grade III embryos. Stem Cells Dev. 18, 423–433 (2009).
Venu, P., Chakraborty, S. & Inamdar, M.S. Analysis of long-term culture properties and pluripotent character of two sibling human embryonic stem cell lines derived from discarded embryos In Vitro Cell Dev. Biol. Anim. 46, 200–205 (2010).
Cheng, E.H. et al. Blastocoel volume is related to successful establishment of human embryonic stem cell lines. Reprod. Biomed. Online 17, 436–444 (2008).
Cowan, C.A. et al. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 350, 1353–1356 (2004).
Stephenson, E.L., Braude, P.R. & Mason, C. International community consensus standard for reporting derivation of human embryonic stem cell lines. Regen. Med. 2, 349–362 (2007).
Hovatta, O. et al. A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Hum. Reprod. 18, 1404–1409 (2003).
Inzunza, J. et al. Derivation of human embryonic stem cell lines in serum replacement medium using postnatal human fibroblasts as feeder cells. Stem Cells 23, 544–549 (2005).
Lee, J.B. et al. Establishment and maintenance of human embryonic stem cell lines on human feeder cells derived from uterine endometrium under serum-free condition. Biol. Reprod. 72, 42–49 (2005).
Richards, M., Fong, C.Y., Chan, W.K., Wong, P.C. & Bongso, A. Human feeders support prolonged undifferentiated growth of human inner cell masses and embryonic stem cells. Nat. Biotechnol. 20, 933–936 (2002).
Crook, J.M. et al. The generation of six clinical-grade human embryonic stem cell lines. Cell Stem Cell 1, 490–494 (2007).
Fu, X. et al. Establishment of clinically compliant human embryonic stem cells in an autologous feeder-free system. Tissue Eng. Part C Methods 17, 927–937 (2011).
Fragouli, E. & Wells, D. Aneuploidy in the human blastocyst. Cytogenet. Genome Res. 133, 149–159 (2011).
Acknowledgements
This work was supported by the Flemish Foundation for Scientific Research (FWO-Vlaanderen (grant no. FWO-3G062910) and by the Concerted Research Actions funding from Bijzonder Onderzoeksfonds to P.D.S. (grant no. BOF GOA 01G01112). S.M.C.S.L. was supported by the Netherlands organization of Scientific Research (NWO) (no. ASPASIA 015.007.037) and the Interuniversity Attraction Poles (PAI) (no. P6/20). P.D.S. is the holder of a fundamental clinical research mandate by the Flemish Foundation for Scientific Research (FWO-Vlaanderen).
Author information
Authors and Affiliations
Contributions
T.O., B.H., P.D.S. and S.M.C.d.S.L. designed the study; T.O., S.L., M.V.d.J. and G.D. performed the experiments; and T.O., B.H., S.L. and S.M.C.d.S.L. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
O'Leary, T., Heindryckx, B., Lierman, S. et al. Derivation of human embryonic stem cells using a post–inner cell mass intermediate. Nat Protoc 8, 254–264 (2013). https://doi.org/10.1038/nprot.2012.157
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.157
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.