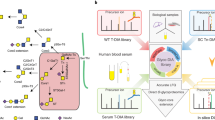
Abstract
Mass spectrometry–based targeted proteomics is a rapidly expanding method for quantifying proteins in complex clinical samples such as plasma. In conjunction with the stable isotope dilution method, selected reaction monitoring (SRM) assays provide unparalleled sensitivity and selectivity for detection and quantification. A crucial factor for robust SRM assays is the reduction of interference by lowering the background. This can be achieved by the selective isolation of a subproteome, such as N-glycosylated proteins, from the original sample. The present protocol includes the development and optimization of SRM assays associated with each peptide of interest and the qualification of assays in the biological matrix to establish the limits of detection and quantification. The protocol also describes the enrichment of formerly N-glycosylated peptides relying on periodate oxidation of glycan moieties attached to the proteins, their immobilization on solid supports through hydrazide chemistry, proteolysis and enzymatic release of the formerly N-glycosylated peptides.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Surinova, S. et al. On the development of plasma protein biomarkers. J. Proteome Res. 10, 5–16 (2011).
Anderson, N.L. & Anderson, N.G. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 (2002).
Gallien, S., Duriez, E. & Domon, B. Selected reaction monitoring applied to proteomics. J. Mass Spectrom. 46, 298–312 (2011).
Picotti, P. et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 (2010).
Hager, J.W. & Yves Le Blanc, J.C. Product ion scanning using a Q-q-Q linear ion trap (Q TRAP) mass spectrometer. Rapid Commun. Mass Spectrom. 17, 1056–1064 (2003).
Hoke, S.H. et al. Transformations in pharmaceutical research and development, driven by innovations in multidimensional mass spectrometry-based technologies. Int. J. Mass Spectrom. 212, 135–196 (2001).
Keshishian, H., Addona, T., Burgess, M., Kuhn, E. & Carr, S.A. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 (2007).
Pan, S. et al. Mass spectrometry based targeted protein quantification: methods and applications. J. Proteome Res. 8, 787–797 (2009).
Desiderio, D.M. & Kai, M. Preparation of stable isotope-incorporated peptide internal standards for field desorption mass spectrometry quantification of peptides in biologic tissue. Biomed. Mass Spectrom. 10, 471–479 (1983).
Anderson, L. & Hunter, C.L. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 (2006).
Kiyonami, R. et al. Increased selectivity, analytical precision, and throughput in targeted proteomics. Mol. Cell. Proteomics 10, 1–11 (2011).
Barnidge, D.R., Goodmanson, M.K., Klee, G.G. & Muddiman, D.C. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-Ms/MS using protein cleavage and isotope dilution mass spectrometry. J. Proteome Res. 3, 644–652 (2004).
Domon, B. Glycosylation as means of reducing sample complexity to enable quantitative proteomics. Proteomics 9, 1488–1491 (2009).
Giron, P., Dayon, L. & Sanchez, J.C. Cysteine tagging for MS-based proteomics. Mass Spectrom. Rev. 30, 366–395.
Zhang, H., Li, X.J., Martin, D.B. & Aebersold, R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 (2003).
Zhang, H. et al. Mass spectrometric detection of tissue proteins in plasma. Mol. Cell. Proteomics 6, 64–71 (2007).
Tian, Y., Zhou, Y., Elliott, S., Aebersold, R. & Zhang, H. Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2, 334–339 (2007).
Bobbitt, J.M. Periodate oxidation of carbohydrates. Adv. Carbohydr. Chem. 48, 1–41 (1956).
Ossola, R. et al. Biomarker validation in blood specimens by selected reaction monitoring mass spectrometry of N-glycosites. Methods Mol. Biol. 728, 179–194 (2011).
Stahl-Zeng, J. et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 6, 1809–1817 (2007).
Chelius, D. & Shaler, T.A. Capture of peptides with N-terminal serine and threonine: a sequence-specific chemical method for peptide mixture simplification. Bioconjug. Chem. 14, 205–211 (2003).
Chen, R. et al. Development of glycoprotein capture based label-free method for the high-throughput screening of differential glycoproteins in hepatocellular carcinoma. Mol. Cell. Proteomics 10, M110.006445 (2011).
Kettenbach, A.N., Rush, J. & Gerber, S.A. Absolute quantification of protein and post-translational modification abundance with stable isotope-labeled synthetic peptides. Nat. Protoc. 6, 175–186 (2011).
Jones, P. et al. PRIDE: new developments and new datasets. Nucleic Acids Res. 36, D878–D883 (2008).
Fusaro, V.A., Mani, D.R., Mesirov, J.P. & Carr, S.A. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 27, 190–198 (2009).
Mallick, P. et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 (2007).
de Graaf, E.L., Altelaar, A.F., van Breukelen, B., Mohammed, S. & Heck, A.J. Improving SRM assay development: a global comparison between triple quadrupole, ion trap, and higher energy CID peptide fragmentation spectra. J. Proteome Res. 10, 4334–4341 (2011).
Prakash, A. et al. Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J. Proteome Res. 8, 2733–2739 (2009).
Cham Mead, J.A., Bianco, L. & Bessant, C. Free computational resources for designing selected reaction monitoring transitions. Proteomics 10, 1106–1126 (2010).
Acknowledgements
This work is supported by FNR (Luxembourg Fonds National de la Recherche) via a PEARL grant and by the Ministry of Higher Education and Research of Luxembourg via the PPM program. We thank S.-Y. Kim for bioinformatics help.
Author information
Authors and Affiliations
Contributions
Y.J.K. designed the experiments, analyzed data and wrote the manuscript. Z.Z.-A. performed the N-glycopeptide enrichment experiment. S.G. performed the SRM assay development, quantitative data analyses and prepared the manuscript. B.D. directed and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Number of N-X-S/T tryptic peptides. (PPT 86 kb)
Supplementary Table 2
Set of reference peptides for retention time correction. (PPT 118 kb)
Supplementary Figure 1
MS/MS spectra of isotopically labeled synthetic peptide, AYLLPAPPAPGnASESEEDR* (n = asparagine with deamidation, R* = arginine with 13C/15N label. (a) Peptide fragmentation using collision induced dissociation (CID) in ion trap. (b) Peptide fragmentation using higher energy collision dissociation (HCD). (PPT 94 kb)
Rights and permissions
About this article
Cite this article
Kim, Y., Zaidi-Ainouch, Z., Gallien, S. et al. Mass spectrometry–based detection and quantification of plasma glycoproteins using selective reaction monitoring. Nat Protoc 7, 859–871 (2012). https://doi.org/10.1038/nprot.2012.023
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.023
This article is cited by
-
A peptide N-terminal protection strategy for comprehensive glycoproteome analysis using hydrazide chemistry based method
Scientific Reports (2015)
-
Quantitation of Permethylated N-Glycans through Multiple-Reaction Monitoring (MRM) LC-MS/MS
Journal of the American Society for Mass Spectrometry (2015)
-
Applications of Multiple Reaction Monitoring to Clinical Glycomics
Chromatographia (2015)
-
Mass spectrometry-based N-glycoproteomics for cancer biomarker discovery
Clinical Proteomics (2014)
-
In-depth analysis of site-specific N-glycosylation in vitronectin from human plasma by tandem mass spectrometry with immunoprecipitation
Analytical and Bioanalytical Chemistry (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.