Abstract
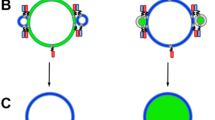
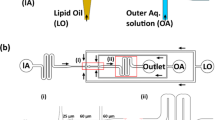
Many biological processes rely on membrane fusion, and therefore assays to study its mechanisms are necessary. Here we report an assay with sensitivity to single-vesicle, and even to single-molecule events using fluorescently labeled vesicle-associated v-SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) liposomes and target-membrane-associated t-SNARE–reconstituted planar, supported bilayers (t-SBLs). Docking and fusion events can be detected using conventional far-field epifluorescence or total internal reflection fluorescence microscopy. In this assay, fusion is dependent on SNAP-25, one of the t-SNARE subunits that is required for fusion in vivo. The success of the assay is due to the use of: (i) bilayers covered with a thin layer of poly(ethylene glycol) (PEG) to control bilayer-bilayer and bilayer-substrate interactions, and (ii) microfluidic flow channels that present many advantages, such as the removal of nonspecifically bound liposomes by flow. The protocol takes 6–8 d to complete. Analysis can take up to 2 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Weber, T. et al. SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772 (1998).
Sudhof, T.C. & Rothman, J.E. Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477 (2009).
Yoon, T.Y., Okumus, B., Zhang, F., Shin, Y.K. & Ha, T. Multiple intermediates in SNARE-induced membrane fusion. Proc. Natl. Acad. Sci. USA 103, 19731–19736 (2006).
Kyoung, M. et al. In vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc. Natl. Acad. Sci. USA 108, E304–E313 (2011).
Diao, J. et al. A single-vesicle content mixing assay for SNARE-mediated membrane fusion. Nat. Commun. 1, 54 (2010).
Smith, E.A. & Weisshaar, J.C. Docking, not fusion, as the rate-limiting step in a SNARE-driven vesicle fusion assay. Biophys. J. 100, 2141–2150 (2011).
Diao, J. et al. A single vesicle-vesicle fusion assay for in vitro studies of SNAREs and accessory proteins. Nat. Protoc. 7, 921–934 (2012).
Karatekin, E. et al. A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc. Natl. Acad. Sci. USA 107, 3517–3521 (2010).
Smith, M.B. et al. Interactive, computer-assisted tracking of speckle trajectories in fluorescence microscopy: application to actin polymerization and membrane fusion. Biophys. J. 101, 1794–1804 (2011).
Brian, A.A. & McConnell, H.M. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc. Natl. Acad. Sci. USA 81, 6159–6163 (1984).
Tamm, L.K. & McConnell, H.M. Supported phospholipid bilayers. Biophys. J. 47, 105–113 (1985).
Fix, M. et al. Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl. Acad. Sci. USA 101, 7311–7316 (2004).
Bowen, M.E., Weninger, K., Brunger, A.T. & Chu, S. Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs). Biophys. J. 87, 3569–3584 (2004).
Liu, T., Tucker, W.C., Bhalla, A., Chapman, E.R. & Weisshaar, J.C. SNARE-driven, 25-millisecond vesicle fusion in vitro. Biophys. J. 89, 2458–2472 (2005).
de Gennes, P.G. Polymers at an interface; a simplified view. Adv. Colloid Interface Sci. 27, 189–209 (1987).
Kenworthy, A.K., Hristova, K., Needham, D. & McIntosh, T.J. Range and magnitude of the steric pressure between bilayers containing phospholipids with covalently attached poly(ethylene glycol). Biophys. J. 68, 1921–1936 (1995).
Milner, S.T. Polymer brushes. Science 251, 905–914 (1991).
Israelachvili, J. The different faces of poly(ethylene glycol). Proc. Natl. Acad. Sci. USA 94, 8378–8379 (1997).
Oesterhelt, F., Rief, M. & Gaub, H.E. Single molecule force spectroscopy by AFM indicates helical structure of poly(ethylene-glycol) in water. N. J. Phys. 1, 6.1–6.11 (1999).
Perrret, E., Leung, A., Morel, A., Feracci, H. & Nassoy, P. Versatile decoration of glass surfaces to probe individual protein-protein interactions and cellular adhesion. Langmuir 18, 846–854 (2002).
Knoll, W. et al. Solid supported lipid membranes: new concepts for the biomimetic functionalization of solid surfaces. Biointerphases 3, Fa125–Fa135 (2008).
Hiergeist, C. & Lipowsky, R. Elastic properties of polymer-decorated membranes. J. De Physique II 6, 1465–1481 (1996).
Kenworthy, A.K., Simon, S.A. & McIntosh, T.J. Structure and phase behavior of lipid suspensions containing phospholipids with covalently attached poly(ethylene glycol). Biophys. J. 68, 1903–1920 (1995).
Lasic, D.D. & Needham, D. The ''Stealth'' liposome: a prototypical biomaterial. Chem. Rev. 95, 2601–2628 (1995).
Albertorio, F. et al. Fluid and air-stable lipopolymer membranes for biosensor applications. Langmuir 21, 7476–7482 (2005).
Tanaka, M. & Sackmann, E. Polymer-supported membranes as models of the cell surface. Nature 437, 656–663 (2005).
Deng, Y. et al. Fluidic and air-stable supported lipid bilayer and cell-mimicking microarrays. J. Am. Chem. Soc. 130, 6267–6271 (2008).
Lin, J., Szymanski, J., Searson, P.C. & Hristova, K. Effect of a polymer cushion on the electrical properties and stability of surface-supported lipid bilayers. Langmuir 26, 3544–3548 (2010).
Wong, J.Y. et al. Polymer-cushioned bilayers. I. A structural study of various preparation methods using neutron reflectometry. Biophys. J. 77, 1445–1457 (1999).
Cornell, B.A. et al. A biosensor that uses ion-channel switches. Nature 387, 580–583 (1997).
Deverall, M.A. et al. Transbilayer coupling of obstructed lipid diffusion in polymer-tethered phospholipid bilayers. Soft Matter 4, 1899–1908 (2008).
Floyd, D.L., Ragains, J.R., Skehel, J.J., Harrison, S.C. & van Oijen, A.M. Single-particle kinetics of influenza virus membrane fusion. Proc. Natl. Acad. Sci. USA 105, 15382–15387 (2008).
Kataoka-Hamai, C., Higuchi, M., Iwai, H. & Miyahara, Y. Detergent-mediated formation of polymer-supported phospholipid bilayers. Langmuir 26, 14600–14605 (2010).
Daniel, S., Albertorio, F. & Cremer, P.S. Making lipid membranes rough, tough, and ready to hit the road. MRS Bull. 31, 536–540 (2006).
Diaz, A.J., Albertorio, F., Daniel, S. & Cremer, P.S. Double cushions preserve transmembrane protein mobility in supported bilayer systems. Langmuir 24, 6820–6826 (2008).
Fasshauer, D., Otto, H., Eliason, W.K., Jahn, R. & Brunger, A.T. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J. Biol. Chem. 272, 28036–28041 (1997).
Hazzard, J., Sudhof, T.C. & Rizo, J. NMR analysis of the structure of synaptobrevin and of its interaction with syntaxin. J. Biomol. NMR 14, 203–207 (1999).
Bright, J.N., Woolf, T.B. & Hoh, J.H. Predicting properties of intrinsically unstructured proteins. Prog. Biophys. Mol. Biol. 76, 131–173 (2001).
Quinn, P., Griffiths, G. & Warren, G. Den sity of newly synthesized plasma membrane proteins in intracellular membranes II. Biochemical studies. J. Cell Biol. 98, 2142–2147 (1984).
Wessels, L., Elting, M.W., Scimeca, D. & Weninger, K. Rapid membrane fusion of individual virus particles with supported lipid bilayers. Biophys. J. 93, 526–538 (2007).
Brunger, A.T., Weninger, K., Bowen, M. & Chu, S. Single-molecule studies of the neuronal SNARE fusion machinery. Annu. Rev. Biochem. 78, 903–928 (2009).
Domanska, M.K., Kiessling, V., Stein, A., Fasshauer, D. & Tamm, L.K. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J. Biol. Chem. 284, 32158–32166 (2009).
Pobbati, A.V., Stein, A. & Fasshauer, D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science 313, 673–676 (2006).
Needham, D. & Nunn, R.S. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys. J. 58, 997–1009 (1990).
Rawicz, W., Smith, B.A., McIntosh, T.J., Simon, S.A. & Evans, E. Elasticity, strength, and water permeability of bilayers that contain raft microdomain-forming lipids. Biophys. J. 94, 4725–4736 (2008).
Nikolaus, J., Stockl, M., Langosch, D., Volkmer, R. & Herrmann, A. Direct visualization of large and protein-free hemifusion diaphragms. Biophys. J. 98, 1192–1199 (2010).
Ohki, S. A mechanism of divalent ion-induced phosphatidylserine membrane fusion. Biochim. Biophys. Acta 689, 1–11 (1982).
Berquand, A. et al. Two-step formation of streptavidin-supported lipid bilayers by PEG-triggered vesicle fusion. Fluorescence and atomic force microscopy characterization. Langmuir 19, 1700–1707 (2003).
Israelachvili, J.N. Intermolecular and Surface Forces (Academic Press, 1991).
Finkelstein, A. Bilayers: formation, measurements, and incorporation of components. Methods Enzymol. 32, 489–501 (1974).
Scott, B.L. et al. Liposome fusion assay to monitor intracellular membrane fusion machines. Methods Enzymol. 372, 274–300 (2003).
Wang, T., Smith, E.A., Chapman, E.R. & Weisshaar, J.C. Lipid mixing and content release in single-vesicle, SNARE-driven fusion assay with 1–5 ms resolution. Biophys. J. 96, 4122–4131 (2009).
Soumpasis, D.M. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 41, 95–97 (1983).
Acknowledgements
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and by a US National Institutes of Health grant to J.E.R. E.K. is indebted to M. Seagar and members of his laboratory (Institut National de la Santé et de la Recherche Médicale (INSERM) UMR641) for introducing him to proteoliposomes, and J. Coleman (Yale University) for teaching him how to express and purify SNARE proteins. We thank J.-P. Henry, F. Darchen and B. Gasnier (Laboratory of Membrane Dynamics and Neurological Diseases, CNRS/Université Paris Descartes UMR 8192, formerly CNRS UPR 1929); T. Melia and A. Gohlke (Department of Cell Biology, Yale University); M. Power (School of Engineering and Applied Science clean room, Yale University); B. O'Shaughnessy and J. Warner at Columbia University for many useful discussions and suggestions; the CNRS for granting a leave of absence to E.K.; and A. Gohlke for help with some of the photos. We thank A. Gohlke, W. Xu, B. Antonny, G. Melikyan, B. O'Shaughnessy, J. Warner and J. Diao for carefully reading and commenting on the manuscript.
Author information
Authors and Affiliations
Contributions
E.K. developed the assay first in the Laboratory of Membrane Dynamics and Neurological Diseases, CNRS/Université Paris Descartes UMR 8192 (formerly CNRS UPR 1929), then in the laboratory of J.E.R. at Yale University. J.E.R. oversaw the project and provided all the material support required since E.K. joined his lab. E.K. wrote the MATLAB analysis programs that are supplied. E.K. wrote the manuscript, which was read and approved by J.E.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Snapshots from a typical movie acquired using TIRFM (256 pixels by 402 pixels, 57 frames/s). (PDF 88 kb)
Supplementary Methods
The supplementary files below are packaged into a single zip file. The files are separated into three folders: (ZIP 67046 kb)
\FRAP\ Three sample FRAP recordings (in Nikon .nd2 file format), Matlab programs, instructions, and auxiliary files.
\Fusion\Epi\ Three sample fusion events cropped from larger movies (10 Hz acquisition rate, far-field epifluorescence mode), sample PointPicker files for analysis of docking and fusion events, Matlab programs, instructions, and auxiliary files. The movies are 16 bit tif stacks that can be opened using e.g. ImageJ.
\Fusion\TIRFM\ Three sample fusion events cropped from larger movies (57 Hz acquisition rate, TIRFM mode), sample SpeckleTrackerJ track files for analysis of docking and fusion events, Matlab programs, instructions, and auxiliary files. The movies are 16 bit tif stacks that can be opened using e.g. ImageJ.
Rights and permissions
About this article
Cite this article
Karatekin, E., Rothman, J. Fusion of single proteoliposomes with planar, cushioned bilayers in microfluidic flow cells. Nat Protoc 7, 903–920 (2012). https://doi.org/10.1038/nprot.2012.019
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.019
This article is cited by
-
A micropatterned substrate for on-surface enzymatic labelling of linearized long DNA molecules
Scientific Reports (2019)
-
Self-assembled vesicles of sodium oleate and chitosan quaternary ammonium salt in acidic or alkaline aqueous solutions
Colloid and Polymer Science (2019)
-
Nanodisc-cell fusion: control of fusion pore nucleation and lifetimes by SNARE protein transmembrane domains
Scientific Reports (2016)
-
Studying calcium-triggered vesicle fusion in a single vesicle-vesicle content and lipid-mixing system
Nature Protocols (2013)
-
A single vesicle-vesicle fusion assay for in vitro studies of SNAREs and accessory proteins
Nature Protocols (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.