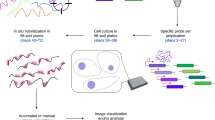
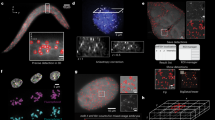
Abstract
Fluorescent in situ hybridization (FISH) allows the quantification of single mRNAs in budding yeast using fluorescently labeled single-stranded DNA probes, a wide-field epifluorescence microscope and a spot-detection algorithm. Fixed yeast cells are attached to coverslips and hybridized with a mixture of FISH probes, each conjugated to several fluorescent dyes. Images of cells are acquired in 3D and maximally projected for single-molecule analysis. Diffraction-limited labeled mRNAs are observed as bright fluorescent spots and can be quantified using a spot-detection algorithm. FISH preserves the spatial distribution of cellular RNA distribution within the cell and the stochastic fluctuations in individual cells that can lead to phenotypic differences within a clonal population. This information, however, is lost if the RNA content is measured on a population of cells by using reverse transcriptase PCR, microarrays or high-throughput sequencing. The FISH procedure and image acquisition described here can be completed in 3 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Singer, R.H. & Ward, D.C. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc. Natl. Acad. Sci. USA 79, 7331–7335 (1982).
Lawrence, J.B. & Singer, R.H. Quantitative analysis of in situ hybridization methods for the detection of actin gene expression. Nucleic Acids Res. 13, 1777–1799 (1985).
Levsky, J.M. & Singer, R.H. Fluorescence in situ hybridization: past, present and future. J. Cell Sci. 116 (Pt 14): 2833–2838 (2003).
Volpi, E.V. & Bridger, J.M. FISH glossary: an overview of the fluorescence in situ hybridization technique. Biotechniques 45, 385–386, 388, 390 passim (2008).
Lawrence, J.B. & Singer, R.H. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45, 407–415 (1986).
Long, R.M. et al. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277, 383–387 (1997).
Gall, J.G. & Pardue, M.L. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc. Natl. Acad. Sci. USA 63, 378–383 (1969).
Kislauskis, E.H. et al. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 123, 165–172 (1993).
Femino, A.M. et al. Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998).
Levsky, J.M. et al. Single-cell gene expression profiling. Science 297, 836–840 (2002).
Gandhi, S.J. et al. Transcription of functionally related constitutive genes is not coordinated. Nat. Struct. Mol. Biol. 18, 27–34 (2011).
Zenklusen, D., Larson, D.R. & Singer, R.H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 (2008).
Silverman, S.J. et al. Metabolic cycling in single yeast cells from unsynchronized steady-state populations limited on glucose or phosphate. Proc. Natl. Acad. Sci. USA 107, 6946–6951 (2010).
Garcia, M. et al. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol. Biol. Cell 18, 362–368 (2007).
Trcek, T. et al. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147, 1484–1497 (2011).
Itzkowitz, S. et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat. Cell Biol. 14, 106–114 (2012).
Thompson, R.E., Larson, D.R. & Webb, W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82, 2775–2783 (2002).
Femino, A.M. et al. Visualization of single molecules of mRNA in situ. Methods Enzymol. 361, 245–304 (2003).
Larson, D.R. et al. Visualization of retrovirus budding with correlated light and electron microscopy. Proc. Natl. Acad. Sci. USA 102, 15453–15458 (2005).
Raj, A. et al. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008).
Raj, A. & Tyagi, S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 472, 365–386 (2010).
Youk, H., Raj, A. & van Oudenaarden, A. Imaging single mRNA molecules in yeast. Methods Enzymol. 470, 429–446 (2010).
Bertrand, E. et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998).
Park, H.Y., Buxbaum, A.R. & Singer, R.H. Single mRNA tracking in live cells. Methods Enzymol. 472, 387–406 (2010).
Tyagi, S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods 6, 331–338 (2009).
Chao, J.A. et al. Structural basis for the coevolution of a viral RNA-protein complex. Nat. Struct. Mol. Biol. 15, 103–105 (2008).
Larson, D.R. et al. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332, 475–478 (2011).
Daigle, N. & Ellenberg, J. λN-GFP: an RNA reporter system for live-cell imaging. Nat. Methods 4, 633–636 (2007).
Takizawa, P.A. & Vale, R.D. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. USA 97, 5273–5278 (2000).
Lange, S. et al. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic 9, 1256–1267 (2008).
Zenklusen, D. & Singer, R.H. Analyzing mRNA expression using single mRNA resolution fluorescent in situ hybridization. Methods Enzymol. 470, 641–659 (2010).
Spellman, P.T. et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273–3297 (1998).
Long, R.M. et al. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 19, 6592–6601 (2000).
Acknowledgements
We thank the Singer laboratory members for critical review of this manuscript. This work was supported by US National Institutes of Health grants GM57071 and GM86217 (awarded to R.H.S.) and a grant from the Canadian Institutes of Health Research (awarded to D.Z.)
Author information
Authors and Affiliations
Contributions
T.T. designed and generated the figures in the paper. T.T., J.A.C., D.Z. contributed to optimization of the multiprobe FISH protocol. D.R.L. wrote the detection algorithm Localize. S.M.S. wrote the cell segmentation plug-in for ImageJ. T.T., J.A.C., D.R.L., H.Y.P., D.Z. and R.H.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
R.H.S. is a consultant with Biosearch Technologies.
Rights and permissions
About this article
Cite this article
Trcek, T., Chao, J., Larson, D. et al. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nat Protoc 7, 408–419 (2012). https://doi.org/10.1038/nprot.2011.451
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.451
This article is cited by
-
Label-free detection of ssDNA base insertion and deletion mutations by surface-enhanced Raman spectroscopy
Analytical and Bioanalytical Chemistry (2022)
-
Tracing DNA paths and RNA profiles in cultured cells and tissues with ORCA
Nature Protocols (2021)
-
Single-molecule fluorescence in-situ hybridization reveals that human SHANK3 mRNA expression varies during development and in autism-associated SHANK3 heterozygosity
Stem Cell Research & Therapy (2018)
-
Rapid and Efficient FISH using Pre-Labeled Oligomer Probes
Scientific Reports (2018)
-
Transcriptional timing and noise of yeast cell cycle regulators—a single cell and single molecule approach
npj Systems Biology and Applications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.