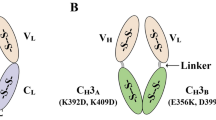
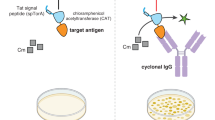
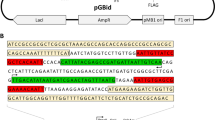
Abstract
Here we describe protocols for the expression of human antibody fragments in Escherichia coli. Antigen-specific clones are identified by soluble fragment ELISA and concentrated by periplasmic preparation. They are then further purified by affinity chromatography. This article provides an overview of expression and purification strategies for human antibody fragments, as well as detailed protocols for the identification of high-affinity binders and for affinity maturation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Choy, E.H. et al. Efficacy of a novel PEGylated humanized anti-TNF fragment (CDP870) in patients with rheumatoid arthritis: a phase II double-blinded, randomized, dose-escalating trial. Rheumatology (Oxford) 41, 1133–1137 (2002).
Cabilly, S. et al. Generation of antibody activity from immunoglobulin polypeptide chains produced in Escherichia coli. Proc. Natl. Acad. Sci. USA 81, 3273–3277 (1984).
Skerra, A. & Pluckthun, A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 240, 1038–1041 (1988).
Wulfing, C. & Pluckthun, A. Protein folding in the periplasm of Escherichia coli. Mol. Microbiol. 12, 685–692 (1994).
Power, B.E. et al. High-level temperature-induced synthesis of an antibody VH-domain in Escherichia coli using the PelB secretion signal. Gene 113, 95–99 (1992).
Steiner, D., Forrer, P., Stumpp, M.T. & Pluckthun, A. Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nat. Biotechnol. 24, 823–831 (2006).
Thie, H., Schirrmann, T., Paschke, M., Dubel, S. & Hust, M. SRP and Sec pathway leader peptides for antibody phage display and antibody fragment production in E. coli. Nat. Biotechnol. 25, 49–54 (2008).
Buchner, J., Brinkmann, U. & Pastan, I. Renaturation of a single-chain immunotoxin facilitated by chaperones and protein disulfide isomerase. Biotechnology (NY) 10, 682–685 (1992).
Lilie, H., Lang, K., Rudolph, R. & Buchner, J. Prolyl isomerases catalyze antibody folding in vitro. Protein Sci. 2, 1490–1496 (1993).
Lowe, D. et al. Aggregation, stability, and formulation of human antibody therapeutics. Adv. Protein Chem. Struct. Biol. 84, 41–61 (2011).
Rothlisberger, D., Honegger, A. & Pluckthun, A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J. Mol. Biol. 347, 773–789 (2005).
Baneyx, F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10, 411–421 (1999).
Lo, B.K.C. Antibody Engineering: Methods and Protocols (Humana Press, 2004).
Jansson, B., Uhlen, M. & Nygren, P.A. All individual domains of staphylococcal protein A show Fab binding. FEMS Immunol. Med. Microbiol. 20, 69–78 (1998).
Bjorck, L. Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J. Immunol. 140, 1194–1197 (1988).
Jonsson, U. et al. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. Biotechniques 11, 620–627 (1991).
Stenberg, E., Persson, B., Roos, H. & Urbaniczky, C. Quantitative-determination of surface concentration of protein with surface-plasmon resonance using radiolabeled proteins. J. Colloid Interf. Sci. 143, 513–526 (1991).
Peters, W.B., Frasca, V. & Brown, R.K. Recent developments in isothermal titration calorimetry label free screening. Comb. Chem. High Throughput Screen 12, 772–790 (2009).
Godber, B. et al. Direct quantification of analyte concentration by resonant acoustic profiling. Clin. Chem. 51, 1962–1972 (2005).
Schnerr, H.R. Lead identification and optimization in crude samples using label free resonant acoustic profiling. J. Mol. Recognit. 23, 597–603 (2010).
Abdiche, Y., Malashock, D., Pinkerton, A. & Pons, J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 377, 209–217 (2008).
Hawkins, R.E., Russell, S.J. & Winter, G. Selection of phage antibodies by binding affinity. Mimicking affinity maturation. J. Mol. Biol. 226, 889–896 (1992).
Lee, C.M., Iorno, N., Sierro, F. & Christ, D. Selection of human antibody fragments by phage display. Nat. Protoc. 2, 3001–3008 (2007).
Sambrook, J. & Russell, D.W. The Condensed Protocols from Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2006).
Jager, M., Gehrig, P. & Pluckthun, A. The scFv fragment of the antibody hu4D5-8: evidence for early premature domain interaction in refolding. J. Mol. Biol. 305, 1111–1129 (2001).
Christ, D., Famm, K. & Winter, G. Repertoires of aggregation-resistant human antibody domains. Protein Eng. Des. Sel. 20, 413–416 (2007).
Dudgeon, K., Famm, K. & Christ, D. Sequence determinants of protein aggregation in human VH domains. Protein Eng. Des. Sel. 22, 217–220 (2009).
Famm, K., Hansen, L., Christ, D. & Winter, G. Thermodynamically stable aggregation-resistant antibody domains through directed evolution. J. Mol. Biol. 376, 926–931 (2008).
Miroux, B. & Walker, J.E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996).
Acknowledgements
Protocols are based on methods originally developed by D. Christ and others in G. Winter's group at the UK Medical Resarch Council (MRC) Laboratory of Molecular Biology. These were further modified at the Garvan Institute of Medical Research and at MedImmune. This work was funded by the Garvan Institute of Medical Research; MedImmune; the Australian National Health and Medical Council; the Australian Research Council; the Cancer Institute, NSW; and the UK MRC.
Author information
Authors and Affiliations
Contributions
D. Lowe, K.D., B.R., P.S., D. Langley, J.A. and P.W. wrote the paper. R.R. wrote the paper and generated figures. L.J. and D.C. wrote the paper and supervised research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Rouet, R., Lowe, D., Dudgeon, K. et al. Expression of high-affinity human antibody fragments in bacteria. Nat Protoc 7, 364–373 (2012). https://doi.org/10.1038/nprot.2011.448
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.448
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.