Abstract
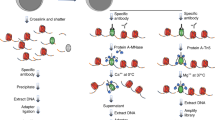
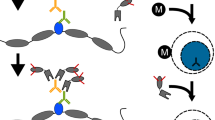
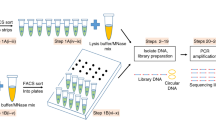
Linear amplification of DNA (LinDA) by T7 polymerase is a versatile and robust method for generating sufficient amounts of DNA for genome-wide studies with minute amounts of cells. LinDA can be coupled to a great number of global profiling technologies. Indeed, chromatin immunoprecipitation coupled to massive parallel sequencing (ChIP-seq) has been achieved for transcription factors and epigenetic modification of chromatin histones with 1,000 to 5,000 cells. LinDA largely simplifies reChIP-seq experiments to monitor co-binding at chromatin target sites. The single-tube design of LinDA is ideal for handling ultrasmall amounts of DNA (<30 pg) and is compatible with automation. The actual hands-on working time is less than 6 h with one overnight reaction. The present protocol describes all materials and critical steps, and provides examples and controls for LinDA. Applications of LinDA for genome-wide analyses of biobank samples and for the study of chromatin conformation and nuclear architecture are in progress.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hurtado, A., Holmes, K.A., Ross-Innes, C.S., Schmidt, D. & Carroll, J.S. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33 (2011).
Ceschin, D.G. et al. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 25, 1132–1146 (2011).
Bau, D. et al. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat. Struct. Mol. Biol. 18, 107–114 (2011).
Park, P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669–680 (2009).
Laird, P.W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 11, 191–203 (2010).
Hah, N. et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634 (2011).
Ozsolak, F. & Milos, P.M. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98 (2011).
Sims, R.J. III et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332, 99–103 (2011).
Wang, Z., Gerstein, M. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009).
Mamanova, L. et al. FRT-seq: amplification-free, strand-specific transcriptome sequencing. Nat. Methods 7, 130–132 (2010).
Ingolia, N.T. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 470, 119–142 (2010).
Guo, H., Ingolia, N.T., Weissman, J.S. & Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010).
Adli, M., Zhu, J. & Bernstein, B.E. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat. Methods 7, 615–618 (2010).
Shankaranarayanan, P. et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat. Methods 8, 565–567 (2011).
Hoeijmakers, W.A., Bartfai, R., Francoijs, K.J. & Stunnenberg, H.G. Linear amplification for deep sequencing. Nat. Protoc. 6, 1026–1036 (2011).
Liu, C.L., Bernstein, B.E. & Schreiber, S.L. Whole genome amplification by T7-based linear amplification of DNA (TLAD): overview. Cold Spring Harb. Protoc. doi:10.1101/pdb.top42 (2008).
Liu, C.L., Schreiber, S.L. & Bernstein, B.E. Development and validation of a T7-based linear amplification for genomic DNA. BMC Genomics 4, 19 (2003).
Zhao, H., Hastie, T., Whitfield, M.L., Borresen-Dale, A.L. & Jeffrey, S.S. Optimization and evaluation of T7-based RNA linear amplification protocols for cDNA microarray analysis. BMC Genomics 3, 31 (2002).
van Bakel, H. et al. Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic Acids Res. 36, e21 (2008).
Xu, Y., Grindley, N.D. & Joyce, C.M. Coordination between the polymerase and 5′-nuclease components of DNA polymerase I of Escherichia coli. J. Biol. Chem. 275, 20949–20955 (2000).
Walker, G.T. Empirical aspects of strand displacement amplification. PCR Methods Appl. 3, 1–6 (1993).
Rong, M., Durbin, R.K. & McAllister, W.T. Template strand switching by T7 RNA polymerase. J. Biol. Chem. 273, 10253–10260 (1998).
Hurtado, A. et al. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature 456, 663–666 (2008).
Mendoza-Parra, M.A., Shankaranarayanan, P. & Gronemeyer, H. Sequential chromatin immunoprecipitation protocol for global analysis through massive parallel sequencing (reChIP-seq). Protocol Exchange, doi:10.1038/protex.2011.1256 (2011).
Fullwood, M.J. et al. An oestrogen receptor-α–bound human chromatin interactome. Nature 462, 58–64 (2009).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Fanelli, M. et al. Pathology tissue-chromatin immunoprecipitation, coupled with high-throughput sequencing, allows the epigenetic profiling of patient samples. Proc. Natl. Acad. Sci. USA 107, 21535–21540 (2010).
Adli, M. & Bernstein, B.E. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat. Protoc. 6, 1656–1668 (2011).
Spits, C. et al. Whole-genome multiple displacement amplification from single cells. Nat. Protoc. 1, 1965–1970 (2006).
Rodrigue, S. et al. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE 4, e6864 (2009).
Kozarewa, I. et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods 6, 291–295 (2009).
Bath, A.J., Milsom, S.E., Gormley, N.A. & Halford, S.E. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 277, 4024–4033 (2002).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Cheung, M.S., Down, T.A., Latorre, I. & Ahringer, J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res. 39, e103 (2011).
Dohm, J.C., Lottaz, C., Borodina, T. & Himmelbauer, H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 36, e105 (2008).
Nelson, J.R. et al. TempliPhi, phi29 DNA polymerase based rolling circle amplification of templates for DNA sequencing. Biotechniques (suppl.) 44–47 (2002).
Hosono, S. et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 13, 954–964 (2003).
Acknowledgements
W.v.G. is supported by the Fondation pour la Recherche Médicale (aide aux projets innovants). This work was supported by funds from the Ligue National Contre le Cancer (laboratoire labelisé), the Institut National du Cancer (INCa) and the European Community contracts LSHC-CT-2005-518417 'EPITRON', LSHG-CT2005-018882 'X-TRA-NET', LSHM-CT2005-018652 'CRESCENDO' and HEALTH-F4-2009-221952 'ATLAS'.
Author information
Authors and Affiliations
Contributions
P.S., L.M.T. and H.G. designed and optimized LinDA. M.-A.M.-P. performed the F9 experiments and did the bioinformatics analysis together with P.S. and W.v.G.; H.G., P.S., M.-A.M.-P. and L.M.T. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
H.G., P.S. and L.M.T. have filed a patent application describing the methods presented in this study (EP11305531.3).
Supplementary information
Supplementary Fig. 1
Multidimensional ChIP-seq comparison including ChIPs done with very small numbers of cells. Four representative screenshots of ERα (purple) and RNA polymerase II (blue) profiles established from H3396 breast cancer cells2, compared with the corresponding H3K4me3 profiles (red; cumulative profiles of two biological replicates). These profiles were established by standard ChIP-seq procedure. Below the corresponding profiles derived LinDA-ChIP-seq assays done with 10,000 and 1,000 cells are displayed (black). Note the coherence between the profiles derived from 10,000 cells with the standard procedure, which is only partially maintained when 1,000 cells were used. (PDF 159 kb)
Supplementary Table 1
Quantification of yields of RNA and double-stranded DNA obtained from representative ChIPs performed with different numbers of cells and two different antibodies. LinDA was performed on ChIPs and total RNA was quantified after the in vitro transcription step; the total DNA was quantified after the final step just before Illumina sequencing. No LinDA was performed on the 2 million cell sample for ERα and the 1 million cells sample for the H3K4me3. The RNA and DNA amounts shown here for the ERα experiments correspond to the same ChIPs for which the sequencing data have been detailed in the Suppl. Table 1 of ref. 14. Note that these are independent ChIP experiments and that numbers cannot be extrapolated linearly. However, the LinDA data are representative and indicate an "apparent experimental amplification factor" between about 2,000-fold (for 5,000 cells, ERα) and 400-fold (for 100,000 cells, ERα (PDF 137 kb)
Supplementary Table 2
The antibody to cell ratios for ChIPs with small number of cells. ChIPs were performed with antibodies directed to ERa (sc-543, Santa Cruz Biotechnology) or H3K4me3 (AB8580, Abcam) using estrogen-treated H3396 breast cancer cells and the standard ChIP protocol. (PDF 124 kb)
Rights and permissions
About this article
Cite this article
Shankaranarayanan, P., Mendoza-Parra, MA., van Gool, W. et al. Single-tube linear DNA amplification for genome-wide studies using a few thousand cells. Nat Protoc 7, 328–339 (2012). https://doi.org/10.1038/nprot.2011.447
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.447
This article is cited by
-
AutoRELACS: automated generation and analysis of ultra-parallel ChIP-seq
Scientific Reports (2020)
-
FoxH1 represses miR-430 during early embryonic development of zebrafish via non-canonical regulation
BMC Biology (2019)
-
LaminA/C regulates epigenetic and chromatin architecture changes upon aging of hematopoietic stem cells
Genome Biology (2018)
-
H3K4me3 epigenomic landscape derived from ChIP-Seq of 1 000 mouse early embryonic cells
Cell Research (2015)
-
GATA-dependent transcriptional and epigenetic control of cardiac lineage specification and differentiation
Cellular and Molecular Life Sciences (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.