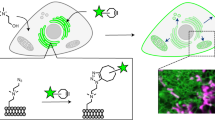
Abstract
It is now recognized that lipids and proteins in cellular membranes are not homogenously distributed. A high degree of membrane order is the biophysical hallmark of cholesterol-enriched lipid rafts, which may induce the lateral sorting of proteins within the membrane. Here we describe a quantitative fluorescence microscopy technique for imaging localized lipid environments and measuring membrane lipid order in live and fixed cells, as well as in intact tissues. The method is based on the spectral ratiometric imaging of the polarity-sensitive membrane dyes Laurdan and di-4-ANEPPDHQ. Laurdan typically requires multiphoton excitation, making it suitable for the imaging of tissues such as whole, living zebrafish embryos, whereas di-4-ANEPPDHQ imaging can be achieved with standard confocal microscopes. This approach, which takes around 4 h, directly examines the organization of cellular membranes and is distinct from alternative approaches that infer membrane order by measuring probe partitioning or dynamics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Simons, K. et al. Functional rafts in cell membranes. Nature 387, 569–572 (1997).
Pike, L.J. Rafts defined: a report on the Keystone symposium on lipid rafts and cell function. J. Lipid Res. 47, 1597–1598 (2006).
Tanimura, N. et al. Dynamic changes in the mobility of LAT in aggregated lipid rafts upon T cell activation. J. Cell Biol. 160, 125–135 (2003).
Owen, D.M. et al. Quantitative microscopy: protein dynamics and membrane organisation. Traffic 10, 962–971 (2009).
Gaus, K. et al. Condensation of the plasma membrane at the site of T lymphocyte activation. J. Cell Biol. 171, 121–131 (2005).
Owen, D.M. et al. High plasma membrane lipid order imaged at the immunological synapse periphery in live T cells. Mol. Membr. Biol. 27, 178–189 (2010).
Rentero, C. et al. Functional implications of plasma membrane condensation for T cell activation. PLoS ONE 3, e2262 (2008).
Gupta, N. et al. Lipid rafts and B cell signaling. Semin. Cell Dev. Biol. 18, 616–626 (2007).
Gaus, K. et al. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 174, 725–734 (2006).
Barman, S. et al. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J. Virol. 81, 12169–12178 (2007).
del Real, G. et al. Blocking of HIV-1 infection by targeting CD4 to nonraft membrane domains. J. Exp. Med. 196, 293–301 (2002).
Scheiffele, P. et al. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274, 2038–2044 (1999).
Ikonen, E. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13, 470–477 (2001).
Hanzal-Bayer, M.F. et al. Lipid rafts and membrane traffic. FEBS Lett. 581, 2098–2104 (2007).
Weber, G. & Farris, F.J. Synthesis and spectral properties of a hydrophobic fluorescent probe: 6-propionyl-2-(dimethylamino)naphthalene. Biochemistry 18, 3075–3078 (1979).
Parasassi, T. et al. Laurdan and Prodan as polarity-sensitive fluorescent membrane probes. J. Fluoresc. 8, 365–737 (1998).
Viard, M. et al. Laurdan solvatochromism: solvent dielectric relaxation and intramolecular excited-state reaction. Biophys. J. 73, 2221–2234 (1997).
Vincent, M. et al. Nanosecond dynamics of a mimicked membrane-water interface observed by time-resolved stokes shift of LAURDAN. Biophys. J. 88, 4337–4350 (2005).
Parasassi, T. et al. Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence. Biophys. J. 57, 1179–1186 (1990).
Parasassi, T. et al. Two-photon fluorescence microscopy of laurdan generalized polarization domains in model and natural membranes. Biophys. J. 72, 2413–2429 (1997).
Owen, D.M. et al. Imaging membrane lipid order in whole, living vertebrate organisms. Biophys. J. 99, L7–L9 (2010).
Yu, W. et al. Fluorescence generalized polarization of cell membranes: a two-photon scanning microscopy approach. Biophys. J. 70, 626–636 (1996).
Gaus, K. et al. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. USA 100, 15554–15559 (2003).
Gaus, K. et al. Condensation of the plasma membrane at the site of T lymphocyte activation. J. Cell Biol. 171, 121–131 (2005).
Kaiser, H. et al. Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. USA 106, 16645–16650 (2009).
Römer, W. et al. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450, 670–675 (2007).
Römer, W. et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell 140, 540–553 (2010).
Obaid, A.L. et al. Novel naphthylstyryl-pyridinium potentiometric dyes offer advantages for neural network analysis. J. Neurosci. Meth. 134, 179–190 (2004).
Jin, L. et al. Cholesterol-enriched lipid domains can be visualized by di-4-ANEPPDHQ with linear and nonlinear optics. Biophys. J. 89, L04–L06 (2005).
Jin, L. et al. Characterization and application of a new optical probe for membrane lipid domains. Biophys. J. 90, 2563–2575 (2006).
Dinic, J. et al. Laurdan and di-4-ANEPPDHQ do not respond to membrane-inserted peptides and are good probes for lipid packing. Biochim. Biophys. Acta 1808, 298–306 (2011).
Janes, P.W. et al. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J. Cell Biol. 147, 447–461 (1999).
Zhang, R.-G. et al. The three-dimensional crystal structure of cholera toxin. J. Mol. Biol. 251, 563–573 (1995).
Eggeling, C. et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1163 (2009).
Wawrezinieck, L. et al. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 89, 4029–4042 (2005).
Bacia, K. et al. Fluorescence correlation spectroscopy relates rafts in model and native membranes. Biophys. J. 87, 1034–1043 (2004).
Kahya, N. et al. Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J. Biol. Chem. 278, 28109–28115 (2003).
Lommerse, P.H.M. et al. Single-molecule diffusion reveals similar mobility for the Lck, H-Ras, and K-Ras membrane anchors. Biophys. J. 91, 1090–1097 (2006).
Schneckenburger, H. et al. Time-gated total internal reflection fluorescence spectroscopy (TG-TIRFS): application to the membrane marker laurdan. J. Microsc. 211, 30–36 (2003).
Grant, D.M. et al. High speed optically sectioned fluorescence lifetime imaging permits study of live cell signaling events. Opt. Express 15, 15656–15673 (2007).
Miguel, L. et al. Primary human CD4+ T cells have diverse levels of membrane lipid order that correlate with their function. J. Immunol. 186, 3505–3516 (2011).
Kim, H.M. et al. A two-photon fluorescent probe for lipid raft imaging: C-Laurdan. Chembiochem 8, 553–559 (2007).
Kim, H.M. et al. Two-photon fluorescent turn-on probe for lipid rafts in live cell and tissue. J. Am. Chem. Soc. 130, 4246–4247 (2008).
Veatch, S.L. et al. Fluorescent probes alter miscibility phase boundaries in ternary vesicles. J. Phys. Chem. 111, 502–504 (2007).
Gaus, K. et al. Visualizing membrane microdomains by Laurdan 2-photon microscopy. Mol. Membr. Biol. 23, 41–48 (2006).
Westerfield, M. The Zebrafish Book: A Guide For the Laboratory Use of Zebrafish (Danio rerio) 4th edn. (University of Oregon Press, 2000).
Acknowledgements
D.M.O. and K.G. acknowledge funding from the Australian Research Council; K.G. also receives funding from the National Health and Medical Research Council of Australia and the Human Frontier Science Program. C.R. is grateful to the CONSOLIDER-INGENIO fellowship (CSD2009-00016, Ministerio de Innovación, Ciencia y Tecnología).
Author information
Authors and Affiliations
Contributions
D.M.O. developed the di-4-ANEPPDHQ protocol and wrote the manuscript. C.R. wrote the GP analysis macro. D.M.O. and A.M. conducted the experiments presented here. D.M.O., A.M. and A.A-S. developed the zebrafish protocol. K.G. developed the Laurdan protocol and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Data 1
ImageJ macro for processing two channel Laurdan or di-4-ANEPPDHQ into GP images. The macro imports TIFF images from two different spectral windows. Using the data flow shown in Figure 6, it generates GP images and pseudo-coloured GP-intensity merged images. (TXT 16 kb)
Rights and permissions
About this article
Cite this article
Owen, D., Rentero, C., Magenau, A. et al. Quantitative imaging of membrane lipid order in cells and organisms. Nat Protoc 7, 24–35 (2012). https://doi.org/10.1038/nprot.2011.419
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.419
This article is cited by
-
Single-molecule localization microscopy reveals STING clustering at the trans-Golgi network through palmitoylation-dependent accumulation of cholesterol
Nature Communications (2024)
-
A new AMPK isoform mediates glucose-restriction induced longevity non-cell autonomously by promoting membrane fluidity
Nature Communications (2023)
-
Lipid saturation controls nuclear envelope function
Nature Cell Biology (2023)
-
Cell membrane fluidity and ROS resistance define DMSO tolerance of cryopreserved synovial MSCs and HUVECs
Stem Cell Research & Therapy (2022)
-
Ceramide-rich microdomains facilitate nuclear envelope budding for non-conventional exosome formation
Nature Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.